| 1 |
|
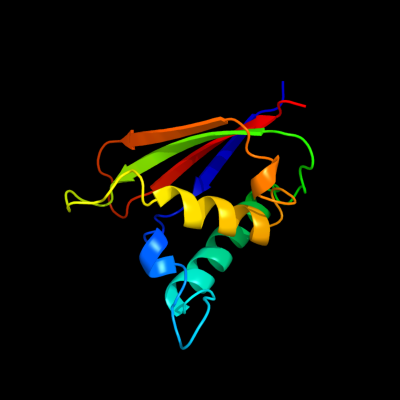
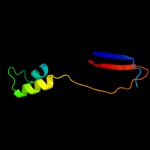
PDB 1q8r chain A
Region: 1 - 118
Aligned: 118
Modelled: 118
Confidence: 100.0%
Identity: 100%
Fold: Bacillus chorismate mutase-like
Superfamily: Holliday junction resolvase RusA
Family: Holliday junction resolvase RusA
Phyre2
| 2 |
|
PDB 1xrs chain B domain 2
Region: 3 - 11
Aligned: 9
Modelled: 9
Confidence: 18.0%
Identity: 44%
Fold: Dodecin subunit-like
Superfamily: D-lysine 5,6-aminomutase beta subunit KamE, N-terminal domain
Family: D-lysine 5,6-aminomutase beta subunit KamE, N-terminal domain
Phyre2
| 3 |
|
PDB 1s1h chain E
Region: 71 - 85
Aligned: 15
Modelled: 15
Confidence: 13.2%
Identity: 47%
PDB header:ribosome
Chain: E: PDB Molecule:40s ribosomal protein s2;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
Phyre2
| 4 |
|
PDB 2az0 chain A domain 1
Region: 64 - 81
Aligned: 18
Modelled: 18
Confidence: 12.5%
Identity: 50%
Fold: ROP-like
Superfamily: FHV B2 protein-like
Family: FHV B2 protein-like
Phyre2
| 5 |
|
PDB 1pkp chain A domain 1
Region: 73 - 85
Aligned: 13
Modelled: 13
Confidence: 12.3%
Identity: 38%
Fold: Ribosomal protein S5 domain 2-like
Superfamily: Ribosomal protein S5 domain 2-like
Family: Translational machinery components
Phyre2
| 6 |
|
PDB 2zkq chain E
Region: 71 - 85
Aligned: 15
Modelled: 15
Confidence: 9.2%
Identity: 53%
PDB header:ribosomal protein/rna
Chain: E: PDB Molecule:rna expansion segment es6 part ii;
PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
Phyre2
| 7 |
|
PDB 3emi chain A
Region: 75 - 89
Aligned: 14
Modelled: 15
Confidence: 8.6%
Identity: 43%
PDB header:cell adhesion
Chain: A: PDB Molecule:hia (adhesin);
PDBTitle: crystal structure of hia 307-422 non-adhesive domain
Phyre2
| 8 |
|
PDB 2qal chain E domain 1
Region: 73 - 85
Aligned: 13
Modelled: 13
Confidence: 7.6%
Identity: 31%
Fold: Ribosomal protein S5 domain 2-like
Superfamily: Ribosomal protein S5 domain 2-like
Family: Translational machinery components
Phyre2
| 9 |
|
PDB 2vnv chain C
Region: 83 - 115
Aligned: 33
Modelled: 33
Confidence: 7.2%
Identity: 9%
PDB header:sugar-binding protein
Chain: C: PDB Molecule:bcla;
PDBTitle: crystal structure of bcla lectin from burkholderia2 cenocepacia in complex with alpha-methyl-mannoside at 1.73 angstrom resolution
Phyre2
| 10 |
|
PDB 1w91 chain A domain 1
Region: 2 - 61
Aligned: 60
Modelled: 60
Confidence: 7.1%
Identity: 12%
Fold: Glycosyl hydrolase domain
Superfamily: Glycosyl hydrolase domain
Family: Composite domain of glycosyl hydrolase families 5, 30, 39 and 51
Phyre2
| 11 |
|
PDB 2xzm chain E
Region: 71 - 83
Aligned: 13
Modelled: 13
Confidence: 6.9%
Identity: 38%
PDB header:ribosome
Chain: E: PDB Molecule:ribosomal protein s5 containing protein;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
Phyre2
| 12 |
|
PDB 2uub chain E domain 1
Region: 73 - 83
Aligned: 11
Modelled: 11
Confidence: 6.2%
Identity: 36%
Fold: Ribosomal protein S5 domain 2-like
Superfamily: Ribosomal protein S5 domain 2-like
Family: Translational machinery components
Phyre2