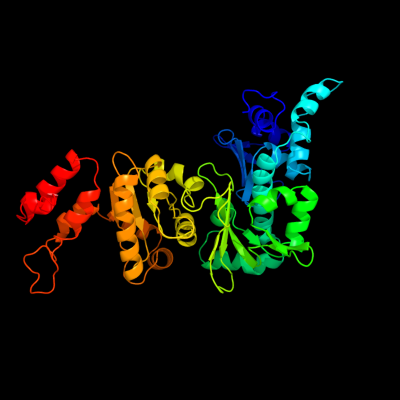
| 1 | c2au3A_
|
|
 |
100.0 |
34 |
PDB header:transferase
Chain: A: PDB Molecule:dna primase;
PDBTitle: crystal structure of the aquifex aeolicus primase (zinc binding and2 rna polymerase domains)
|
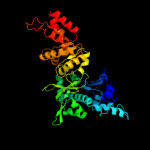
| 2 | d1dd9a_
|
|
 |
100.0 |
99 |
Fold:DNA primase core
Superfamily:DNA primase core
Family:DNA primase DnaG catalytic core |
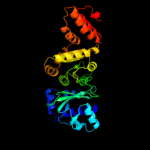
| 3 | c1dd9A_
|
|
 |
100.0 |
99 |
PDB header:transferase
Chain: A: PDB Molecule:dna primase;
PDBTitle: structure of the dnag catalytic core
|
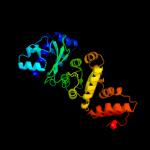
| 4 | c1nuiA_
|
|
 |
100.0 |
20 |
PDB header:replication
Chain: A: PDB Molecule:dna primase/helicase;
PDBTitle: crystal structure of the primase fragment of bacteriophage t7 primase-2 helicase protein
|
| 5 | d1d0qa_
|
|
 |
100.0 |
48 |
Fold:Rubredoxin-like
Superfamily:Zinc beta-ribbon
Family:DNA primase zinc finger |
| 6 | c1q57G_
|
|
 |
99.9 |
23 |
PDB header:transferase
Chain: G: PDB Molecule:dna primase/helicase;
PDBTitle: the crystal structure of the bifunctional primase-helicase of2 bacteriophage t7
|
| 7 | d1nuia1
|
|
 |
99.9 |
20 |
Fold:DNA primase core
Superfamily:DNA primase core
Family:Primase fragment of primase-helicase protein |
| 8 | d1t6t1_
|
|
 |
99.7 |
23 |
Fold:Toprim domain
Superfamily:Toprim domain
Family:Toprim domain |
| 9 | d1t3wa_
|
|
 |
99.2 |
98 |
Fold:DNA primase DnaG, C-terminal domain
Superfamily:DNA primase DnaG, C-terminal domain
Family:DNA primase DnaG, C-terminal domain |
| 10 | d2fcja1
|
|
 |
98.8 |
20 |
Fold:Toprim domain
Superfamily:Toprim domain
Family:Toprim domain |
| 11 | c2r6cG_
|
|
 |
98.2 |
14 |
PDB header:replication
Chain: G: PDB Molecule:dnag primase, helicase binding domain;
PDBTitle: crystal form bh2
|
| 12 | c2r5uD_
|
|
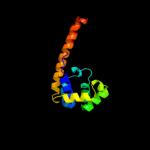 |
96.4 |
8 |
PDB header:hydrolase
Chain: D: PDB Molecule:replicative dna helicase;
PDBTitle: crystal structure of the n-terminal domain of dnab helicase from2 mycobacterium tuberculosis
|
| 13 | c2q6tB_
|
|
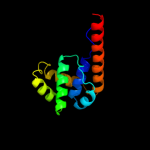 |
96.2 |
7 |
PDB header:hydrolase
Chain: B: PDB Molecule:dnab replication fork helicase;
PDBTitle: crystal structure of the thermus aquaticus dnab monomer
|
| 14 | d1jwea_
|
|
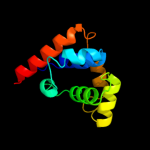 |
95.3 |
11 |
Fold:N-terminal domain of DnaB helicase
Superfamily:N-terminal domain of DnaB helicase
Family:N-terminal domain of DnaB helicase |
| 15 | c1vddC_
|
|
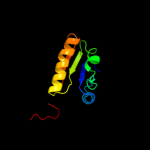 |
93.8 |
19 |
PDB header:recombination
Chain: C: PDB Molecule:recombination protein recr;
PDBTitle: crystal structure of recombinational repair protein recr
|
| 16 | d1vdda_
|
|
 |
93.4 |
19 |
Fold:Recombination protein RecR
Superfamily:Recombination protein RecR
Family:Recombination protein RecR |
| 17 | c3gxvA_
|
|
 |
92.8 |
8 |
PDB header:hydrolase/replication
Chain: A: PDB Molecule:replicative dna helicase;
PDBTitle: three-dimensional structure of n-terminal domain of dnab2 helicase from helicobacter pylori and its interactions with3 primase
|
| 18 | d1b79a_
|
|
 |
92.6 |
11 |
Fold:N-terminal domain of DnaB helicase
Superfamily:N-terminal domain of DnaB helicase
Family:N-terminal domain of DnaB helicase |
| 19 | c2e9hA_
|
|
 |
91.7 |
29 |
PDB header:translation
Chain: A: PDB Molecule:eukaryotic translation initiation factor 5;
PDBTitle: solution structure of the eif-5_eif-2b domain from human2 eukaryotic translation initiation factor 5
|
| 20 | c3bgwD_
|
|
 |
90.5 |
8 |
PDB header:replication
Chain: D: PDB Molecule:dnab-like replicative helicase;
PDBTitle: the structure of a dnab-like replicative helicase and its interactions2 with primase
|
| 21 | c1neeA_ |
|
not modelled |
86.8 |
25 |
PDB header:translation
Chain: A: PDB Molecule:probable translation initiation factor 2 beta
PDBTitle: structure of archaeal translation factor aif2beta from2 methanobacterium thermoautrophicum
|
| 22 | c2vyeA_ |
|
not modelled |
86.7 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:replicative dna helicase;
PDBTitle: crystal structure of the dnac-ssdna complex
|
| 23 | c2dcuB_ |
|
not modelled |
75.6 |
25 |
PDB header:translation
Chain: B: PDB Molecule:translation initiation factor 2 beta subunit;
PDBTitle: crystal structure of translation initiation factor aif2betagamma2 heterodimer with gdp
|
| 24 | c4a17Y_ |
|
not modelled |
73.4 |
32 |
PDB header:ribosome
Chain: Y: PDB Molecule:rpl37a;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 2.
|
| 25 | c2pfsA_ |
|
not modelled |
71.4 |
6 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:universal stress protein;
PDBTitle: crystal structure of universal stress protein from nitrosomonas2 europaea
|
| 26 | c3nmeA_ |
|
not modelled |
71.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:sex4 glucan phosphatase;
PDBTitle: structure of a plant phosphatase
|
| 27 | c2imgA_ |
|
not modelled |
68.4 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:dual specificity protein phosphatase 23;
PDBTitle: crystal structure of dual specificity protein phosphatase2 23 from homo sapiens in complex with ligand malate ion
|
| 28 | c3cc4Z_ |
|
not modelled |
67.6 |
22 |
PDB header:ribosome
Chain: Z: PDB Molecule:50s ribosomal protein l37ae;
PDBTitle: co-crystal structure of anisomycin bound to the 50s ribosomal subunit
|
| 29 | d1v3aa_ |
|
not modelled |
67.5 |
13 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Dual specificity phosphatase-like |
| 30 | c2oudA_ |
|
not modelled |
67.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:dual specificity protein phosphatase 10;
PDBTitle: crystal structure of the catalytic domain of human mkp5
|
| 31 | c3emuA_ |
|
not modelled |
66.6 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:leucine rich repeat and phosphatase domain
PDBTitle: crystal structure of a leucine rich repeat and phosphatase2 domain containing protein from entamoeba histolytica
|
| 32 | c3k7aM_ |
|
not modelled |
63.8 |
23 |
PDB header:transcription
Chain: M: PDB Molecule:transcription initiation factor iib;
PDBTitle: crystal structure of an rna polymerase ii-tfiib complex
|
| 33 | c2qa4Z_ |
|
not modelled |
63.6 |
21 |
PDB header:ribosome
Chain: Z: PDB Molecule:50s ribosomal protein l37ae;
PDBTitle: a more complete structure of the the l7/l12 stalk of the2 haloarcula marismortui 50s large ribosomal subunit
|
| 34 | c1yshD_ |
|
not modelled |
63.6 |
36 |
PDB header:structural protein/rna
Chain: D: PDB Molecule:ribosomal protein l37a;
PDBTitle: localization and dynamic behavior of ribosomal protein l30e
|
| 35 | d1nuia2 |
|
not modelled |
61.4 |
14 |
Fold:Rubredoxin-like
Superfamily:Zinc beta-ribbon
Family:DNA primase zinc finger |
| 36 | c2esbA_ |
|
not modelled |
61.4 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:dual specificity protein phosphatase 18;
PDBTitle: crystal structure of human dusp18
|
| 37 | c1oheA_ |
|
not modelled |
61.3 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:cdc14b2 phosphatase;
PDBTitle: structure of cdc14b phosphatase with a peptide ligand
|
| 38 | d1ffkw_ |
|
not modelled |
60.9 |
25 |
Fold:Rubredoxin-like
Superfamily:Zn-binding ribosomal proteins
Family:Ribosomal protein L37ae |
| 39 | c3rgqA_ |
|
not modelled |
59.5 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein-tyrosine phosphatase mitochondrial 1;
PDBTitle: crystal structure of ptpmt1 in complex with pi(5)p
|
| 40 | d1ohea2 |
|
not modelled |
59.2 |
15 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Dual specificity phosphatase-like |
| 41 | d1vqoz1 |
|
not modelled |
59.0 |
21 |
Fold:Rubredoxin-like
Superfamily:Zn-binding ribosomal proteins
Family:Ribosomal protein L37ae |
| 42 | d1wiia_ |
|
not modelled |
58.5 |
29 |
Fold:Rubredoxin-like
Superfamily:Zinc beta-ribbon
Family:Putative zinc binding domain |
| 43 | c2zkrz_ |
|
not modelled |
57.8 |
21 |
PDB header:ribosomal protein/rna
Chain: Z: PDB Molecule:e site t-rna;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 44 | d1jj2y_ |
|
not modelled |
56.5 |
25 |
Fold:Rubredoxin-like
Superfamily:Zn-binding ribosomal proteins
Family:Ribosomal protein L37ae |
| 45 | c2r0bA_ |
|
not modelled |
56.1 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:serine/threonine/tyrosine-interacting protein;
PDBTitle: crystal structure of human tyrosine phosphatase-like2 serine/threonine/tyrosine-interacting protein
|
| 46 | c1yn9B_ |
|
not modelled |
55.3 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:polynucleotide 5'-phosphatase;
PDBTitle: crystal structure of baculovirus rna 5'-phosphatase2 complexed with phosphate
|
| 47 | c2y96A_ |
|
not modelled |
54.6 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:dual specificity phosphatase dupd1;
PDBTitle: structure of human dual-specificity phosphatase 27
|
| 48 | c2gwoC_ |
|
not modelled |
54.0 |
26 |
PDB header:hydrolase
Chain: C: PDB Molecule:dual specificity protein phosphatase 13;
PDBTitle: crystal structure of tmdp
|
| 49 | c3jyw9_ |
|
not modelled |
53.4 |
36 |
PDB header:ribosome
Chain: 9: PDB Molecule:60s ribosomal protein l43;
PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
|
| 50 | c3rz2B_ |
|
not modelled |
51.7 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:protein tyrosine phosphatase type iva 1;
PDBTitle: crystal of prl-1 complexed with peptide
|
| 51 | d1vhra_ |
|
not modelled |
50.6 |
20 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Dual specificity phosphatase-like |
| 52 | c2g6zB_ |
|
not modelled |
50.2 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:dual specificity protein phosphatase 5;
PDBTitle: crystal structure of human dusp5
|
| 53 | c2i50A_ |
|
not modelled |
49.7 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:ubiquitin carboxyl-terminal hydrolase 16;
PDBTitle: solution structure of ubp-m znf-ubp domain
|
| 54 | d1i9sa_ |
|
not modelled |
49.7 |
22 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Dual specificity phosphatase-like |
| 55 | c2c46B_ |
|
not modelled |
49.5 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:mrna capping enzyme;
PDBTitle: crystal structure of the human rna guanylyltransferase and2 5'-phosphatase
|
| 56 | c3cw2M_ |
|
not modelled |
49.5 |
22 |
PDB header:translation
Chain: M: PDB Molecule:translation initiation factor 2 subunit beta;
PDBTitle: crystal structure of the intact archaeal translation2 initiation factor 2 from sulfolobus solfataricus .
|
| 57 | d1mkpa_ |
|
not modelled |
49.0 |
14 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Dual specificity phosphatase-like |
| 58 | c1s1i9_ |
|
not modelled |
48.5 |
29 |
PDB header:ribosome
Chain: 9: PDB Molecule:60s ribosomal protein l43;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
|
| 59 | c3fldA_ |
|
not modelled |
47.7 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein trai;
PDBTitle: crystal structure of the trai c-terminal domain
|
| 60 | c3s4oB_ |
|
not modelled |
46.2 |
17 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:protein tyrosine phosphatase-like protein;
PDBTitle: protein tyrosine phosphatase (putative) from leishmania major
|
| 61 | d2z3va1 |
|
not modelled |
46.2 |
14 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 62 | c2k8dA_ |
|
not modelled |
45.6 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:peptide methionine sulfoxide reductase msrb;
PDBTitle: solution structure of a zinc-binding methionine sulfoxide reductase
|
| 63 | d1rxda_ |
|
not modelled |
45.3 |
15 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Dual specificity phosphatase-like |
| 64 | c3e90B_ |
|
not modelled |
45.2 |
40 |
PDB header:hydrolase
Chain: B: PDB Molecule:ns3 protease;
PDBTitle: west nile vi rus ns2b-ns3protease in complexed with2 inhibitor naph-kkr-h
|
| 65 | c2e0tA_ |
|
not modelled |
44.9 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:dual specificity phosphatase 26;
PDBTitle: crystal structure of catalytic domain of dual specificity phosphatase2 26, ms0830 from homo sapiens
|
| 66 | d2ijob1 |
|
not modelled |
44.9 |
40 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Viral proteases |
| 67 | d1xria_ |
|
not modelled |
44.9 |
23 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Dual specificity phosphatase-like |
| 68 | c1yz4A_ |
|
not modelled |
44.8 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:dual specificity phosphatase-like 15 isoform a;
PDBTitle: crystal structure of dusp15
|
| 69 | d1muma_ |
|
not modelled |
44.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 70 | d1befa_ |
|
not modelled |
44.3 |
36 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Viral proteases |
| 71 | c2i6oA_ |
|
not modelled |
44.3 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:sulfolobus solfataricus protein tyrosine
PDBTitle: crystal structure of the complex of the archaeal sulfolobus2 ptp-fold phosphatase with phosphopeptides n-g-(p)y-k-n
|
| 72 | c2nt2C_ |
|
not modelled |
42.6 |
11 |
PDB header:hydrolase
Chain: C: PDB Molecule:protein phosphatase slingshot homolog 2;
PDBTitle: crystal structure of slingshot phosphatase 2
|
| 73 | c3hcjB_ |
|
not modelled |
41.9 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:peptide methionine sulfoxide reductase;
PDBTitle: structure of msrb from xanthomonas campestris (oxidized2 form)
|
| 74 | d1w0ba_ |
|
not modelled |
41.9 |
20 |
Fold:Spectrin repeat-like
Superfamily:Alpha-hemoglobin stabilizing protein AHSP
Family:Alpha-hemoglobin stabilizing protein AHSP |
| 75 | d2fp7b1 |
|
not modelled |
41.9 |
40 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Viral proteases |
| 76 | d1z8ua1 |
|
not modelled |
40.5 |
20 |
Fold:Spectrin repeat-like
Superfamily:Alpha-hemoglobin stabilizing protein AHSP
Family:Alpha-hemoglobin stabilizing protein AHSP |
| 77 | d1jeya2 |
|
not modelled |
40.2 |
22 |
Fold:vWA-like
Superfamily:vWA-like
Family:Ku70 subunit N-terminal domain |
| 78 | d1pnoa_ |
|
not modelled |
39.5 |
16 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Transhydrogenase domain III (dIII) |
| 79 | c2vdwA_ |
|
not modelled |
39.0 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:vaccinia virus capping enzyme d1 subunit;
PDBTitle: guanosine n7 methyl-transferase sub-complex (d1-d12) of the2 vaccinia virus mrna capping enzyme
|
| 80 | d1d4oa_ |
|
not modelled |
38.5 |
18 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Transhydrogenase domain III (dIII) |
| 81 | c2kxoA_ |
|
not modelled |
37.4 |
7 |
PDB header:cell cycle
Chain: A: PDB Molecule:cell division topological specificity factor;
PDBTitle: solution nmr structure of the cell division regulator mine protein2 from neisseria gonorrhoeae
|
| 82 | c2pdtD_ |
|
not modelled |
37.3 |
25 |
PDB header:circadian clock protein
Chain: D: PDB Molecule:vivid pas protein vvd;
PDBTitle: 2.3 angstrom structure of phosphodiesterase treated vivid
|
| 83 | c3lyeA_ |
|
not modelled |
36.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:oxaloacetate acetyl hydrolase;
PDBTitle: crystal structure of oxaloacetate acetylhydrolase
|
| 84 | d1twfj_ |
|
not modelled |
36.0 |
27 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:RNA polymerase subunit RPB10
Family:RNA polymerase subunit RPB10 |
| 85 | d1dl6a_ |
|
not modelled |
36.0 |
25 |
Fold:Rubredoxin-like
Superfamily:Zinc beta-ribbon
Family:Transcriptional factor domain |
| 86 | c1pt9B_ |
|
not modelled |
35.7 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nad(p) transhydrogenase, mitochondrial;
PDBTitle: crystal structure analysis of the diii component of transhydrogenase2 with a thio-nicotinamide nucleotide analogue
|
| 87 | c1zzwA_ |
|
not modelled |
35.4 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:dual specificity protein phosphatase 10;
PDBTitle: crystal structure of catalytic domain of human map kinase2 phosphatase 5
|
| 88 | d2fomb1 |
|
not modelled |
35.4 |
33 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Viral proteases |
| 89 | c2p3qA_ |
|
not modelled |
35.3 |
17 |
PDB header:viral protein,transferase
Chain: A: PDB Molecule:type ii methyltransferase;
PDBTitle: crystal structure of dengue methyltransferase in complex with gpppg2 and s-adenosyl-l-homocysteine
|
| 90 | d1ywfa1 |
|
not modelled |
34.5 |
22 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Mycobacterial PtpB-like |
| 91 | d1g26a_ |
|
not modelled |
34.3 |
50 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Granulin repeat
Family:Granulin repeat |
| 92 | d1jmva_ |
|
not modelled |
34.0 |
7 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 93 | d1xmxa_ |
|
not modelled |
33.8 |
12 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:Hypothetical protein VC1899 |
| 94 | d1ef4a_ |
|
not modelled |
33.7 |
17 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:RNA polymerase subunit RPB10
Family:RNA polymerase subunit RPB10 |
| 95 | c2l1uA_ |
|
not modelled |
33.3 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methionine-r-sulfoxide reductase b2, mitochondrial;
PDBTitle: structure-functional analysis of mammalian msrb2 protein
|
| 96 | c2j17A_ |
|
not modelled |
32.4 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:tyrosine-protein phosphatase yil113w;
PDBTitle: ptyr bound form of sdp-1
|
| 97 | c3e0mB_ |
|
not modelled |
32.3 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:peptide methionine sulfoxide reductase msra/msrb
PDBTitle: crystal structure of fusion protein of msra and msrb
|
| 98 | c1ee8A_ |
|
not modelled |
32.2 |
21 |
PDB header:dna binding protein
Chain: A: PDB Molecule:mutm (fpg) protein;
PDBTitle: crystal structure of mutm (fpg) protein from thermus thermophilus hb8
|
| 99 | d1pfta_ |
|
not modelled |
32.1 |
25 |
Fold:Rubredoxin-like
Superfamily:Zinc beta-ribbon
Family:Transcriptional factor domain |
| 100 | d1l1da_ |
|
not modelled |
31.5 |
9 |
Fold:Mss4-like
Superfamily:Mss4-like
Family:SelR domain |
| 101 | c3gn5B_ |
|
not modelled |
31.2 |
16 |
PDB header:dna binding protein
Chain: B: PDB Molecule:hth-type transcriptional regulator mqsa (ygit/b3021);
PDBTitle: structure of the e. coli protein mqsa (ygit/b3021)
|
| 102 | d2dkta2 |
|
not modelled |
31.2 |
38 |
Fold:Zinc hairpin stack
Superfamily:Zinc hairpin stack
Family:Zinc hairpin stack |
| 103 | c3cezA_ |
|
not modelled |
30.8 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methionine-r-sulfoxide reductase;
PDBTitle: crystal structure of methionine-r-sulfoxide reductase from2 burkholderia pseudomallei
|
| 104 | c2vbcA_ |
|
not modelled |
30.6 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:dengue 4 ns3 full-length protein;
PDBTitle: crystal structure of the ns3 protease-helicase from dengue2 virus
|
| 105 | d1vd4a_ |
|
not modelled |
28.9 |
14 |
Fold:Rubredoxin-like
Superfamily:Zinc beta-ribbon
Family:Transcriptional factor domain |
| 106 | c3r9jD_ |
|
not modelled |
28.8 |
17 |
PDB header:cell cycle,hydrolase/cell cycle
Chain: D: PDB Molecule:cell division topological specificity factor;
PDBTitle: 4.3a resolution structure of a mind-mine(i24n) protein complex
|
| 107 | d2fiya1 |
|
not modelled |
28.7 |
19 |
Fold:FdhE-like
Superfamily:FdhE-like
Family:FdhE-like |
| 108 | c1wrmA_ |
|
not modelled |
28.7 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:dual specificity phosphatase 22;
PDBTitle: crystal structure of jsp-1
|
| 109 | c3idfA_ |
|
not modelled |
28.1 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:usp-like protein;
PDBTitle: the crystal structure of a usp-like protein from wolinella2 succinogenes to 2.0a
|
| 110 | c1drwA_ |
|
not modelled |
27.9 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: escherichia coli dhpr/nhdh complex
|
| 111 | d1di1a_ |
|
not modelled |
27.7 |
28 |
Fold:Terpenoid synthases
Superfamily:Terpenoid synthases
Family:Aristolochene/pentalenene synthase |
| 112 | d1xm0a1 |
|
not modelled |
27.1 |
11 |
Fold:Mss4-like
Superfamily:Mss4-like
Family:SelR domain |
| 113 | d1p32a_ |
|
not modelled |
27.1 |
17 |
Fold:Mitochondrial glycoprotein MAM33-like
Superfamily:Mitochondrial glycoprotein MAM33-like
Family:Mitochondrial glycoprotein MAM33-like |
| 114 | c3lkwA_ |
|
not modelled |
26.9 |
24 |
PDB header:viral protein,hydrolase
Chain: A: PDB Molecule:fusion protein of nonstructural protein 2b and
PDBTitle: crystal structure of dengue virus 1 ns2b/ns3 protease active2 site mutant
|
| 115 | c2ysaA_ |
|
not modelled |
26.9 |
50 |
PDB header:metal binding protein
Chain: A: PDB Molecule:retinoblastoma-binding protein 6;
PDBTitle: solution structure of the zinc finger cchc domain from the2 human retinoblastoma-binding protein 6 (retinoblastoma-3 binding q protein 1, rbq-1)
|
| 116 | d1yqfa1 |
|
not modelled |
26.0 |
33 |
Fold:Mitochondrial glycoprotein MAM33-like
Superfamily:Mitochondrial glycoprotein MAM33-like
Family:Mitochondrial glycoprotein MAM33-like |
| 117 | c2pmzN_ |
|
not modelled |
25.8 |
17 |
PDB header:translation, transferase
Chain: N: PDB Molecule:dna-directed rna polymerase subunit n;
PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
|
| 118 | d1vh4a_ |
|
not modelled |
25.7 |
18 |
Fold:Single-stranded right-handed beta-helix
Superfamily:Stabilizer of iron transporter SufD
Family:Stabilizer of iron transporter SufD |
| 119 | c3qv0A_ |
|
not modelled |
25.4 |
39 |
PDB header:protein binding
Chain: A: PDB Molecule:mitochondrial acidic protein mam33;
PDBTitle: crystal structure of saccharomyces cerevisiae mam33
|
| 120 | c3f62A_ |
|
not modelled |
25.1 |
29 |
PDB header:cytokine
Chain: A: PDB Molecule:interleukin 18 binding protein;
PDBTitle: crystal structure of human il-18 in complex with ectromelia virus il-2 18 binding protein
|