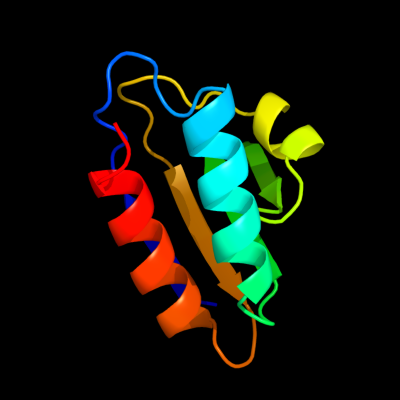
1 d1cm3a_
100.0
98
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like2 d1ka5a_
100.0
33
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like3 c3ihsB_
100.0
37
PDB header: transport proteinChain: B: PDB Molecule: phosphocarrier protein hpr;PDBTitle: crystal structure of a phosphocarrier protein hpr from2 bacillus anthracis str. ames
4 d1mo1a_
100.0
46
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like5 d1ptfa_
100.0
35
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like6 d1qr5a_
100.0
35
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like7 c3le1B_
100.0
43
PDB header: transferaseChain: B: PDB Molecule: phosphotransferase system, hpr-related proteins;PDBTitle: crystal structure of apohpr monomer from thermoanaerobacter2 tengcongensis
8 d1pcha_
100.0
34
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like9 d2hpra_
100.0
36
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like10 d1zvvj1
100.0
45
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like11 d2nzul1
100.0
40
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like12 d1vq3a_
63.6
32
Fold: PurS-likeSuperfamily: PurS-likeFamily: PurS subunit of FGAM synthetase13 c2dgbA_
60.6
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein purs;PDBTitle: structure of thermus thermophilus purs in the p21 form
14 c2jpiA_
58.0
14
PDB header: structural genomicsChain: A: PDB Molecule: hypothetical protein;PDBTitle: chemical shift assignments of pa4090 from pseudomonas2 aeruginosa
15 d1gtda_
50.4
10
Fold: PurS-likeSuperfamily: PurS-likeFamily: PurS subunit of FGAM synthetase16 c2zw2B_
42.4
23
PDB header: ligaseChain: B: PDB Molecule: putative uncharacterized protein sts178;PDBTitle: crystal structure of formylglycinamide ribonucleotide amidotransferase2 iii from sulfolobus tokodaii (stpurs)
17 c3goeA_
42.1
13
PDB header: recombination, replicationChain: A: PDB Molecule: dna repair protein rad60;PDBTitle: molecular mimicry of sumo promotes dna repair
18 c2ekeC_
18.4
13
PDB header: ligase/protein bindingChain: C: PDB Molecule: ubiquitin-like protein smt3;PDBTitle: structure of a sumo-binding-motif mimic bound to smt3p-2 ubc9p: conservation of a noncovalent ubiquitin-like3 protein-e2 complex as a platform for selective4 interactions within a sumo pathway
19 d2etna2
14.9
21
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: GreA transcript cleavage factor, C-terminal domain20 c2etnA_
13.4
21
PDB header: transcriptionChain: A: PDB Molecule: anti-cleavage anti-grea transcription factorPDBTitle: crystal structure of thermus aquaticus gfh1
21 d1t4aa_
not modelled
13.2
13
Fold: PurS-likeSuperfamily: PurS-likeFamily: PurS subunit of FGAM synthetase22 d1yema_
not modelled
12.8
21
Fold: CYTH-like phosphatasesSuperfamily: CYTH-like phosphatasesFamily: CYTH domain23 d1srva_
not modelled
12.6
19
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain24 d1r11a3
not modelled
12.3
17
Fold: MutS N-terminal domain-likeSuperfamily: tRNA-intron endonuclease N-terminal domain-likeFamily: tRNA-intron endonuclease N-terminal domain-like25 c2e6gI_
not modelled
12.2
20
PDB header: hydrolaseChain: I: PDB Molecule: 5'-nucleotidase sure;PDBTitle: crystal structure of the stationary phase survival protein sure from2 thermus thermophilus hb8 in complex with phosphate
26 c2dc4A_
not modelled
11.2
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: 165aa long hypothetical protein;PDBTitle: structure of ph1012 protein from pyrococcus horikoshii ot3
27 d1dk7a_
not modelled
11.2
19
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain28 d2vbua1
not modelled
10.9
31
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin kinase-likeFamily: CTP-dependent riboflavin kinase-like29 c3k6qB_
not modelled
10.1
29
PDB header: ligand binding proteinChain: B: PDB Molecule: putative ligand binding protein;PDBTitle: crystal structure of an antitoxin part of a putative toxin/antitoxin2 system (swol_0700) from syntrophomonas wolfei subsp. wolfei at 1.80 a3 resolution
30 d2f23a2
not modelled
10.0
23
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: GreA transcript cleavage factor, C-terminal domain31 c3bmbB_
not modelled
10.0
20
PDB header: rna binding proteinChain: B: PDB Molecule: regulator of nucleoside diphosphate kinase;PDBTitle: crystal structure of a new rna polymerase interacting2 protein
32 d1we3a2
not modelled
9.5
20
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain33 d1l5xa_
not modelled
9.1
17
Fold: SurE-likeSuperfamily: SurE-likeFamily: SurE-like34 c2pn0D_
not modelled
8.7
26
PDB header: transcriptionChain: D: PDB Molecule: prokaryotic transcription elongation factorPDBTitle: prokaryotic transcription elongation factor grea/greb from2 nitrosomonas europaea
35 d1euvb_
not modelled
8.6
13
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ubiquitin-related36 d1aopa2
not modelled
8.6
24
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: Duplicated SiR/NiR-like domains 1 and 337 d1sjpa2
not modelled
8.6
23
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain38 c3m6cA_
not modelled
8.1
23
PDB header: chaperoneChain: A: PDB Molecule: 60 kda chaperonin 1;PDBTitle: crystal structure of mycobacterium tuberculosis groel1 apical domain
39 c2p4vA_
not modelled
8.1
15
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor greb;PDBTitle: crystal structure of the transcript cleavage factor, greb2 at 2.6a resolution
40 d1w96c1
not modelled
7.8
38
Fold: Barrel-sandwich hybridSuperfamily: Rudiment single hybrid motifFamily: BC C-terminal domain-like41 c2l66B_
not modelled
7.8
13
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, abrb family;PDBTitle: the dna-recognition fold of sso7c4 suggests a new member of spovt-abrb2 superfamily from archaea.
42 c2v4oB_
not modelled
7.6
17
PDB header: hydrolaseChain: B: PDB Molecule: multifunctional protein sur e;PDBTitle: crystal structure of salmonella typhimurium sure at 2.752 angstrom resolution in monoclinic form
43 d1grja2
not modelled
7.6
26
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: GreA transcript cleavage factor, C-terminal domain44 c2jxxA_
not modelled
7.4
14
PDB header: protein bindingChain: A: PDB Molecule: nfatc2-interacting protein;PDBTitle: nmr solution structure of ubiquitin-like domain of2 nfatc2ip. northeast structural genomics consortium target3 hr5627
45 d2fy9a1
not modelled
7.4
12
Fold: Double-split beta-barrelSuperfamily: AbrB/MazE/MraZ-likeFamily: AbrB N-terminal domain-like46 c1ztgD_
not modelled
7.3
7
PDB header: dna, rna binding protein/dnaChain: D: PDB Molecule: poly(rc)-binding protein 1;PDBTitle: human alpha polyc binding protein kh1
47 d1yfba1
not modelled
7.1
29
Fold: Double-split beta-barrelSuperfamily: AbrB/MazE/MraZ-likeFamily: AbrB N-terminal domain-like48 d1kida_
not modelled
6.9
20
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain49 d1b5sa_
not modelled
6.8
33
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: CAT-like50 d1ulza1
not modelled
6.8
13
Fold: Barrel-sandwich hybridSuperfamily: Rudiment single hybrid motifFamily: BC C-terminal domain-like51 d2uyzb1
not modelled
6.6
11
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ubiquitin-related52 c1ibaA_
not modelled
6.5
12
PDB header: phoshphotransferaseChain: A: PDB Molecule: glucose permease;PDBTitle: glucose permease (domain iib), nmr, 11 structures
53 d2io3b1
not modelled
6.3
7
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ubiquitin-related54 c2ro5B_
not modelled
6.2
22
PDB header: transcriptionChain: B: PDB Molecule: stage v sporulation protein t;PDBTitle: rdc-refined solution structure of the n-terminal dna2 recognition domain of the bacillus subtilis transition-3 state regulator spovt
55 c2ii4C_
not modelled
6.2
5
PDB header: transferaseChain: C: PDB Molecule: lipoamide acyltransferase component of branched-chainPDBTitle: crystal structure of a cubic core of the dihydrolipoamide2 acyltransferase (e2b) component in the branched-chain alpha-ketoacid3 dehydrogenase complex (bckdc), coenzyme a-bound form
56 d1dpba_
not modelled
6.2
19
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: CAT-like57 c2yx5A_
not modelled
6.1
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0062 protein mj1593;PDBTitle: crystal structure of methanocaldococcus jannaschii purs, one of the2 subunits of formylglycinamide ribonucleotide amidotransferase in the3 purine biosynthetic pathway
58 d2j9ga1
not modelled
6.1
22
Fold: Barrel-sandwich hybridSuperfamily: Rudiment single hybrid motifFamily: BC C-terminal domain-like59 d1q2ha_
not modelled
6.1
50
Fold: Dimerisation interlockSuperfamily: Phenylalanine zipperFamily: Adapter protein APS, dimerisation domain60 c2w1tB_
not modelled
6.0
24
PDB header: transcriptionChain: B: PDB Molecule: stage v sporulation protein t;PDBTitle: crystal structure of b. subtilis spovt
61 d2bfdb2
not modelled
5.8
21
Fold: TK C-terminal domain-likeSuperfamily: TK C-terminal domain-likeFamily: Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain62 c3gebC_
not modelled
5.6
14
PDB header: hydrolaseChain: C: PDB Molecule: eyes absent homolog 2;PDBTitle: crystal structure of edeya2
63 c2hjwA_
not modelled
5.4
25
PDB header: ligaseChain: A: PDB Molecule: acetyl-coa carboxylase 2;PDBTitle: crystal structure of the bc domain of acc2
64 d1j9ja_
not modelled
5.3
13
Fold: SurE-likeSuperfamily: SurE-likeFamily: SurE-like65 c2k8hA_
not modelled
5.3
13
PDB header: signaling proteinChain: A: PDB Molecule: small ubiquitin protein;PDBTitle: solution structure of sumo from trypanosoma brucei
66 d1w96a1
not modelled
5.1
38
Fold: Barrel-sandwich hybridSuperfamily: Rudiment single hybrid motifFamily: BC C-terminal domain-like67 d1oela2
not modelled
5.1
18
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain