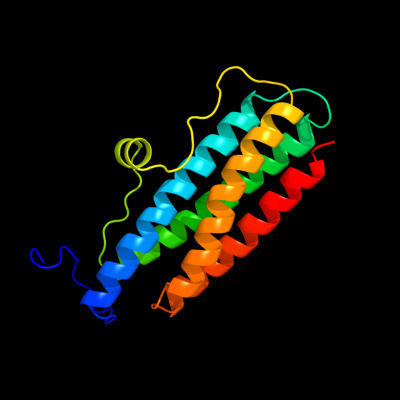
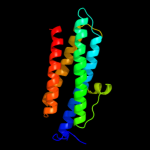
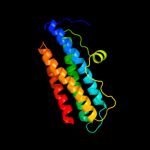
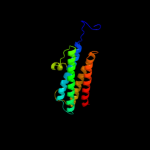
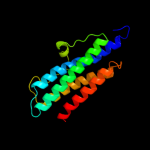
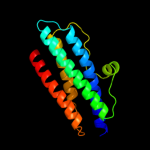
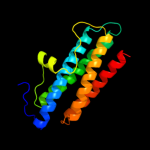
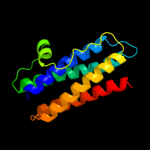
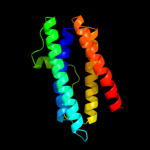
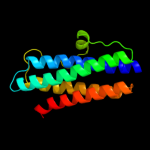
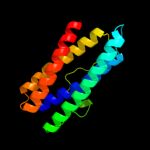
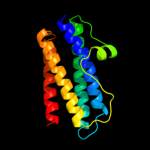
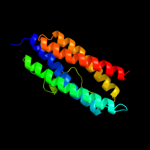
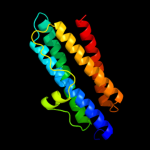
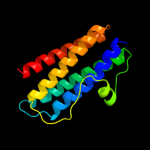
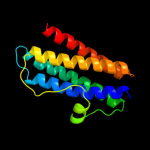
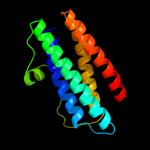
1 d1dpsa_
100.0
99
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin2 c2f7nA_
100.0
25
PDB header: dna binding proteinChain: A: PDB Molecule: dna-binding stress response protein, dps family;PDBTitle: structure of d. radiodurans dps-1
3 d1tjoa_
100.0
22
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin4 d1vela_
100.0
35
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin5 d1o9ra_
100.0
55
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin6 c2z90D_
100.0
28
PDB header: dna binding proteinChain: D: PDB Molecule: starvation-inducible dna-binding protein or finePDBTitle: crystal structure of the second dps from mycobacterium2 smegmatis
7 c2yjkF_
100.0
31
PDB header: metal-binding proteinChain: F: PDB Molecule: afp;PDBTitle: structure of dps from microbacterium arborescens in the2 high iron form
8 d2fjca1
100.0
22
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin9 c2vxxB_
100.0
18
PDB header: dna-binding proteinChain: B: PDB Molecule: starvation induced dna binding protein;PDBTitle: x-ray structure of dpsa from thermosynechococcus elongatus
10 d2yw6a1
100.0
37
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin11 c3kwoA_
100.0
20
PDB header: oxidoreductaseChain: A: PDB Molecule: putative bacterioferritin;PDBTitle: crystal structure of putative bacterioferritin from2 campylobacter jejuni
12 c2c6rA_
100.0
24
PDB header: dna-binding proteinChain: A: PDB Molecule: dna-binding stress response protein, dps family;PDBTitle: fe-soaked crystal structure of the dps92 from deinococcus2 radiodurans
13 d1ji4a_
100.0
24
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin14 c2xgwA_
100.0
26
PDB header: metal binding proteinChain: A: PDB Molecule: peroxide resistance protein;PDBTitle: zinc-bound crystal structure of streptococcus pyogenes dpr
15 d1n1qa_
100.0
28
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin16 c3iq1A_
100.0
19
PDB header: metal transportChain: A: PDB Molecule: dps family protein;PDBTitle: crystal structure of dps protein from vibrio cholerae o1, a member of2 a broad superfamily of ferritin-like diiron-carboxylate proteins
17 c2wlaA_
100.0
26
PDB header: oxidoreductaseChain: A: PDB Molecule: dps-like peroxide resistance protein;PDBTitle: streptococcus pyogenes dpr
18 d1umna_
100.0
26
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin19 d2bk6a1
100.0
25
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin20 c2d5kC_
100.0
24
PDB header: metal binding proteinChain: C: PDB Molecule: dps family protein;PDBTitle: crystal structure of dps from staphylococcus aureus
21 c2pybC_
not modelled
100.0
20
PDB header: metal transportChain: C: PDB Molecule: neutrophil activating protein;PDBTitle: napa protein from borrelia burgdorferi
22 c2c41K_
not modelled
100.0
20
PDB header: iron-binding/oxidation proteinChain: K: PDB Molecule: dps family dna-binding stress response protein;PDBTitle: x-ray structure of dps from thermosynechococcus elongatus
23 c2chpC_
not modelled
100.0
21
PDB header: dna-binding proteinChain: C: PDB Molecule: metalloregulation dna-binding stress protein;PDBTitle: crystal structure of the dodecameric ferritin mrga from2 b.subtilis 168
24 d1ji5a_
not modelled
100.0
22
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin25 d1jiga_
not modelled
100.0
25
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin26 d1zs3a1
not modelled
100.0
10
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin27 d1zuja1
not modelled
100.0
9
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin28 c2clbP_
not modelled
99.9
13
PDB header: metal binding proteinChain: P: PDB Molecule: dps-like protein;PDBTitle: the structure of the dps-like protein from sulfolobus2 solfataricus reveals a bacterioferritin-like di-metal3 binding site within a dps-like dodecameric assembly
29 c2vzbA_
not modelled
99.9
13
PDB header: metal transportChain: A: PDB Molecule: putative bacterioferritin-related protein;PDBTitle: a dodecameric thioferritin in the bacterial domain, characterization2 of the bacterioferritin-related protein from bacteroides fragilis
30 d1nf4a_
not modelled
99.8
16
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin31 d2htna1
not modelled
99.8
15
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin32 c3gvyC_
not modelled
99.8
12
PDB header: metal binding proteinChain: C: PDB Molecule: bacterioferritin;PDBTitle: crystal structure of bacterioferritin from r.sphaeroides
33 c2qqyA_
not modelled
99.8
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: sigma b operon;PDBTitle: crystal structure of ferritin like, diiron-carboxylate proteins from2 bacillus anthracis str. ames
34 c3fvbB_
not modelled
99.8
13
PDB header: metal binding proteinChain: B: PDB Molecule: bacterioferritin;PDBTitle: crystal structure of ferritin (bacterioferritin) from2 brucella melitensis
35 c3bknB_
not modelled
99.8
15
PDB header: metal binding proteinChain: B: PDB Molecule: bacterioferritin;PDBTitle: the structure of mycobacterial bacterioferritin
36 c3r2rA_
not modelled
99.8
18
PDB header: metal binding proteinChain: A: PDB Molecule: bacterioferritin;PDBTitle: 1.65a resolution structure of iron soaked ftna from pseudomonas2 aeruginosa (ph 6.0)
37 d1jgca_
not modelled
99.8
14
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin38 d2fkza1
not modelled
99.8
12
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin39 d1vlga_
not modelled
99.4
14
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin40 d1s3qa1
not modelled
99.4
12
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin41 c3qz3A_
not modelled
99.4
15
PDB header: oxidoreductaseChain: A: PDB Molecule: ferritin;PDBTitle: the crystal structure of ferritin from vibrio cholerae o1 biovar el2 tor str. n16961
42 c3e6sD_
not modelled
99.3
12
PDB header: oxidoreductaseChain: D: PDB Molecule: ferritin;PDBTitle: crystal structure of ferritin soaked with iron from pseudo-2 nitzschia multiseries
43 c2jd8C_
not modelled
99.3
9
PDB header: metal transportChain: C: PDB Molecule: ferritin homolog;PDBTitle: crystal structure of the zn-soaked ferritin from the2 hyperthermophilic archaeal anaerobe pyrococcus furiosus
44 d1euma_
not modelled
99.3
14
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin45 c3bvkC_
not modelled
99.3
11
PDB header: oxidoreductaseChain: C: PDB Molecule: ferritin;PDBTitle: structural basis for the iron uptake mechanism of helicobacter pylori2 ferritin
46 d1krqa_
not modelled
99.3
11
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin47 c3qd8M_
not modelled
99.3
6
PDB header: metal binding proteinChain: M: PDB Molecule: probable bacterioferritin bfrb;PDBTitle: crystal structure of mycobacterium tuberculosis bfrb
48 d1j30a_
not modelled
99.1
13
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin49 d1lkoa1
not modelled
99.1
7
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin50 d1yv1a1
not modelled
99.0
9
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin51 d1lb3a_
not modelled
99.0
12
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin52 d2za7a1
not modelled
99.0
14
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin53 d1yuza1
not modelled
99.0
9
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin54 c3oghB_
not modelled
99.0
5
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: protein ycie;PDBTitle: crystal structure of ycie protein from e. coli cft073, a member of2 ferritine-like superfamily of diiron-containing four-helix-bundle3 proteins
55 d1r03a_
not modelled
98.9
11
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin56 d1mfra_
not modelled
98.9
15
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin57 d2ceia1
not modelled
98.9
11
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin58 d1rcda_
not modelled
98.8
12
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin59 d2oh3a1
not modelled
98.8
12
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: AMB4284-like60 d1nnqa1
not modelled
98.8
12
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin61 d1z6om1
not modelled
98.7
10
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin62 c1dvbA_
not modelled
98.7
7
PDB header: electron transportChain: A: PDB Molecule: rubrerythrin;PDBTitle: rubrerythrin
63 c3a9qR_
not modelled
98.6
11
PDB header: oxidoreductaseChain: R: PDB Molecule: PDBTitle: crystal structure analysis of e173a variant of the soybean2 ferritin sfer4
64 c2hr5B_
not modelled
98.5
12
PDB header: metal binding proteinChain: B: PDB Molecule: rubrerythrin;PDBTitle: pf1283- rubrerythrin from pyrococcus furiosus iron bound form
65 c1yuzB_
not modelled
98.3
10
PDB header: oxidoreductaseChain: B: PDB Molecule: nigerythrin;PDBTitle: partially reduced state of nigerythrin
66 d2fzfa1
not modelled
98.2
13
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin67 d1bg7a_
not modelled
98.0
11
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin68 c2ib0A_
not modelled
98.0
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical alanine rich protein;PDBTitle: crystal structure of a conserved hypothetical protein, rv2844, from2 mycobacterium tuberculosis
69 d2ib0a1
not modelled
98.0
10
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Rv2844-like70 c3qhbA_
not modelled
97.8
12
PDB header: metal transportChain: A: PDB Molecule: symerythrin;PDBTitle: crystal structure of oxidized symerythrin from cyanophora paradoxa
71 d1jkva_
not modelled
97.5
14
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Manganese catalase (T-catalase)72 d1vjxa_
not modelled
97.2
7
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin73 d1z6oa1
not modelled
95.9
12
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin74 c3q4nA_
not modelled
93.5
13
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein mj0754;PDBTitle: crystal structure of hypothetical protein mj0754 from methanococcus2 jannaschii dsm 2661
75 c1u7mB_
not modelled
92.4
20
PDB header: de novo proteinChain: B: PDB Molecule: four-helix bundle model;PDBTitle: solution structure of a diiron protein model: due ferri(ii)2 turn mutant
76 c1lt1G_
not modelled
91.5
23
PDB header: de novo proteinChain: G: PDB Molecule: l13g-df1;PDBTitle: sliding helix induced change of coordination geometry in a2 model di-mn(ii) protein
77 c3hiuB_
not modelled
90.1
11
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: the crystal structure of protein (xcc3681) from xanthomonas2 campestris pv. campestris str. atcc 33913
78 c2hz8A_
not modelled
84.1
11
PDB header: de novo proteinChain: A: PDB Molecule: de novo designed diiron protein;PDBTitle: qm/mm structure refined from nmr-structure of a single2 chain diiron protein
79 d2gyqa1
not modelled
83.6
9
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: YciF-like80 c3fseB_
not modelled
80.8
13
PDB header: hydrolaseChain: B: PDB Molecule: two-domain protein containing dj-1/thij/pfpi-like andPDBTitle: crystal structure of a two-domain protein containing dj-1/thij/pfpi-2 like and ferritin-like domains (ava_4496) from anabaena variabilis3 atcc 29413 at 1.90 a resolution
81 d2gs4a1
not modelled
58.6
6
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: YciF-like82 d2cwla1
not modelled
53.4
13
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Manganese catalase (T-catalase)83 c2bnlE_
not modelled
49.2
35
PDB header: stress-responseChain: E: PDB Molecule: modulator protein rsbr;PDBTitle: the structure of the n-terminal domain of rsbr
84 c2x7lP_
not modelled
27.8
15
PDB header: immune systemChain: P: PDB Molecule: hiv rev;PDBTitle: implications of the hiv-1 rev dimer structure at 3.2a2 resolution for multimeric binding to the rev response3 element
85 c3lphD_
not modelled
26.3
15
PDB header: viral proteinChain: D: PDB Molecule: protein rev;PDBTitle: crystal structure of the hiv-1 rev dimer
86 d1t98a2
not modelled
22.6
17
Fold: STAT-likeSuperfamily: MukF C-terminal domain-likeFamily: MukF C-terminal domain-like87 d2nr5a1
not modelled
22.4
28
Fold: Ferritin-likeSuperfamily: SO2669-likeFamily: SO2669-like88 c2l18A_
not modelled
19.4
9
PDB header: oxidoreductaseChain: A: PDB Molecule: arsenate reductase;PDBTitle: an arsenate reductase in the phosphate binding state
89 d1rxqa_
not modelled
16.8
9
Fold: DinB/YfiT-like putative metalloenzymesSuperfamily: DinB/YfiT-like putative metalloenzymesFamily: YfiT-like putative metal-dependent hydrolases90 c2qf9B_
not modelled
14.5
5
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative secreted protein;PDBTitle: crystal structure of putative secreted protein duf305 from2 streptomyces coelicolor
91 d2hh6a1
not modelled
14.3
11
Fold: Left-handed superhelixSuperfamily: BH3980-likeFamily: BH3980-like92 d2o3la1
not modelled
13.3
8
Fold: Left-handed superhelixSuperfamily: BH3980-likeFamily: BH3980-like93 c1c9lA_
not modelled
11.9
44
PDB header: endocytosis/exocytosisChain: A: PDB Molecule: clathrin;PDBTitle: peptide-in-groove interactions link target proteins to the2 b-propeller of clathrin
94 d1otha1
not modelled
11.5
10
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase95 d1utca2
not modelled
11.3
44
Fold: 7-bladed beta-propellerSuperfamily: Clathrin heavy-chain terminal domainFamily: Clathrin heavy-chain terminal domain96 c2pt7G_
not modelled
10.7
11
PDB header: hydrolase/protein bindingChain: G: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of cag virb11 (hp0525) and an inhibitory protein2 (hp1451)
97 c3ggzH_
not modelled
10.5
28
PDB header: protein transport, endocytosisChain: H: PDB Molecule: vacuolar protein-sorting-associated protein 46;PDBTitle: crystal structure of s.cerevisiae ist1 n-terminal domain in2 complex with did2 mim motif
98 c3ggzE_
not modelled
10.3
28
PDB header: protein transport, endocytosisChain: E: PDB Molecule: vacuolar protein-sorting-associated protein 46;PDBTitle: crystal structure of s.cerevisiae ist1 n-terminal domain in2 complex with did2 mim motif
99 c3ggzF_
not modelled
10.3
28
PDB header: protein transport, endocytosisChain: F: PDB Molecule: vacuolar protein-sorting-associated protein 46;PDBTitle: crystal structure of s.cerevisiae ist1 n-terminal domain in2 complex with did2 mim motif