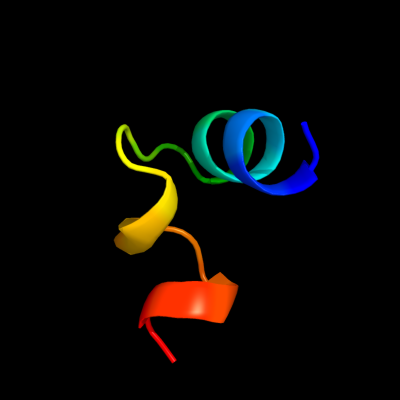| 1 |
|
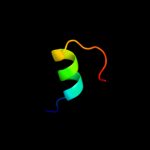
PDB 3hxx chain A
Region: 4 - 26
Aligned: 23
Modelled: 23
Confidence: 24.5%
Identity: 39%
PDB header:ligase
Chain: A: PDB Molecule:alanyl-trna synthetase;
PDBTitle: crystal structure of catalytic fragment of e. coli alars in complex2 with amppcp
Phyre2
| 2 |
|
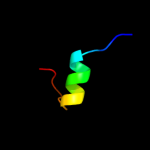
PDB 2fb6 chain A
Region: 40 - 53
Aligned: 14
Modelled: 14
Confidence: 20.6%
Identity: 14%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved hypothetical protein;
PDBTitle: structure of conserved protein of unknown function bt1422 from2 bacteroides thetaiotaomicron
Phyre2
| 3 |
|
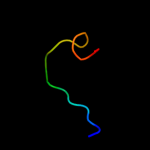
PDB 1yfs chain B
Region: 4 - 26
Aligned: 23
Modelled: 23
Confidence: 19.7%
Identity: 39%
PDB header:ligase
Chain: B: PDB Molecule:alanyl-trna synthetase;
PDBTitle: the crystal structure of alanyl-trna synthetase in complex2 with l-alanine
Phyre2
| 4 |
|
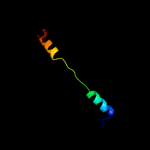
PDB 2zzf chain A
Region: 4 - 23
Aligned: 20
Modelled: 20
Confidence: 12.1%
Identity: 30%
PDB header:ligase
Chain: A: PDB Molecule:alanyl-trna synthetase;
PDBTitle: crystal structure of alanyl-trna synthetase without2 oligomerization domain
Phyre2
| 5 |
|
PDB 3rf1 chain B
Region: 1 - 19
Aligned: 19
Modelled: 19
Confidence: 11.3%
Identity: 42%
PDB header:ligase
Chain: B: PDB Molecule:glycyl-trna synthetase alpha subunit;
PDBTitle: the crystal structure of glycyl-trna synthetase subunit alpha from2 campylobacter jejuni subsp. jejuni nctc 11168
Phyre2
| 6 |
|
PDB 2i3s chain F
Region: 80 - 93
Aligned: 14
Modelled: 14
Confidence: 10.1%
Identity: 50%
PDB header:cell cycle
Chain: F: PDB Molecule:checkpoint serine/threonine-protein kinase;
PDBTitle: bub3 complex with bub1 glebs motif
Phyre2
| 7 |
|
PDB 1j5w chain A
Region: 1 - 19
Aligned: 19
Modelled: 19
Confidence: 9.8%
Identity: 42%
Fold: Class II aaRS and biotin synthetases
Superfamily: Class II aaRS and biotin synthetases
Family: Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain
Phyre2
| 8 |
|
PDB 1ejx chain B
Region: 45 - 79
Aligned: 35
Modelled: 35
Confidence: 9.4%
Identity: 20%
Fold: beta-clip
Superfamily: Urease, beta-subunit
Family: Urease, beta-subunit
Phyre2
| 9 |
|
PDB 2pso chain A domain 1
Region: 52 - 68
Aligned: 17
Modelled: 17
Confidence: 7.7%
Identity: 35%
Fold: TBP-like
Superfamily: Bet v1-like
Family: STAR domain
Phyre2
| 10 |
|
PDB 4ubp chain B
Region: 45 - 79
Aligned: 35
Modelled: 35
Confidence: 7.1%
Identity: 26%
Fold: beta-clip
Superfamily: Urease, beta-subunit
Family: Urease, beta-subunit
Phyre2
| 11 |
|
PDB 1e9y chain A domain 1
Region: 45 - 79
Aligned: 35
Modelled: 35
Confidence: 6.7%
Identity: 20%
Fold: beta-clip
Superfamily: Urease, beta-subunit
Family: Urease, beta-subunit
Phyre2
| 12 |
|
PDB 2k4b chain A
Region: 10 - 49
Aligned: 40
Modelled: 40
Confidence: 6.1%
Identity: 15%
PDB header:dna binding protein
Chain: A: PDB Molecule:transcriptional regulator;
PDBTitle: copr repressor structure
Phyre2
| 13 |
|
PDB 2vn2 chain B
Region: 27 - 58
Aligned: 32
Modelled: 32
Confidence: 6.1%
Identity: 19%
PDB header:replication
Chain: B: PDB Molecule:chromosome replication initiation protein;
PDBTitle: crystal structure of the n-terminal domain of dnad protein2 from geobacillus kaustophilus hta426
Phyre2
| 14 |
|
PDB 1avo chain A
Region: 7 - 42
Aligned: 36
Modelled: 36
Confidence: 5.6%
Identity: 36%
PDB header:proteasome activator
Chain: A: PDB Molecule:11s regulator;
PDBTitle: proteasome activator reg(alpha)
Phyre2
| 15 |
|
PDB 1jb0 chain K
Region: 32 - 48
Aligned: 17
Modelled: 17
Confidence: 5.3%
Identity: 47%
Fold: Photosystem I reaction center subunit X, PsaK
Superfamily: Photosystem I reaction center subunit X, PsaK
Family: Photosystem I reaction center subunit X, PsaK
Phyre2