1 c2h3eB_
100.0
100
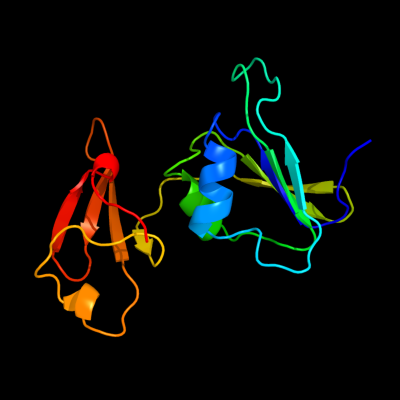
PDB header: transferaseChain: B: PDB Molecule: aspartate carbamoyltransferase regulatory chain;PDBTitle: structure of wild-type e. coli aspartate transcarbamoylase in the2 presence of n-phosphonacetyl-l-isoasparagine at 2.3a resolution
2 c1pg5B_
100.0
34
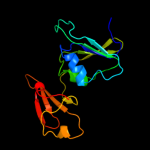
PDB header: transferaseChain: B: PDB Molecule: aspartate carbamoyltransferase regulatory chain;PDBTitle: crystal structure of the unligated (t-state) aspartate2 transcarbamoylase from the extremely thermophilic archaeon sulfolobus3 acidocaldarius
3 c2ywwA_
100.0
35
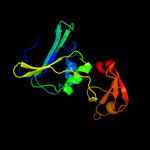
PDB header: metal binding proteinChain: A: PDB Molecule: aspartate carbamoyltransferase regulatory chain;PDBTitle: crystal structure of aspartate carbamoyltransferase2 regulatory chain from methanocaldococcus jannaschii
4 c2be7E_
100.0
57
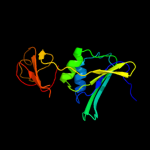
PDB header: transferaseChain: E: PDB Molecule: aspartate carbamoyltransferase regulatory chain;PDBTitle: crystal structure of the unliganded (t-state) aspartate2 transcarbamoylase of the psychrophilic bacterium moritella profunda
5 d2fzcb1
100.0
100
Fold: Ferredoxin-likeSuperfamily: Aspartate carbamoyltransferase, Regulatory-chain, N-terminal domainFamily: Aspartate carbamoyltransferase, Regulatory-chain, N-terminal domain6 d2atcb1
100.0
88
Fold: Ferredoxin-likeSuperfamily: Aspartate carbamoyltransferase, Regulatory-chain, N-terminal domainFamily: Aspartate carbamoyltransferase, Regulatory-chain, N-terminal domain7 d1pg5b1
100.0
42
Fold: Ferredoxin-likeSuperfamily: Aspartate carbamoyltransferase, Regulatory-chain, N-terminal domainFamily: Aspartate carbamoyltransferase, Regulatory-chain, N-terminal domain8 d2fzcb2
99.8
100
Fold: Rubredoxin-likeSuperfamily: Aspartate carbamoyltransferase, Regulatory-chain, C-terminal domainFamily: Aspartate carbamoyltransferase, Regulatory-chain, C-terminal domain9 d2atcb2
99.7
90
Fold: Rubredoxin-likeSuperfamily: Aspartate carbamoyltransferase, Regulatory-chain, C-terminal domainFamily: Aspartate carbamoyltransferase, Regulatory-chain, C-terminal domain10 d1pg5b2
99.7
23
Fold: Rubredoxin-likeSuperfamily: Aspartate carbamoyltransferase, Regulatory-chain, C-terminal domainFamily: Aspartate carbamoyltransferase, Regulatory-chain, C-terminal domain11 c1xrzA_
51.4
50
PDB header: transcriptionChain: A: PDB Molecule: zinc finger y-chromosomal protein;PDBTitle: nmr structure of a zinc finger with cyclohexanylalanine2 substituted for the central aromatic residue
12 c7znfA_
42.9
46
PDB header: zinc finger dna binding domainChain: A: PDB Molecule: zinc finger;PDBTitle: alternating zinc fingers in the human male associated2 protein zfy: 2d nmr structure of an even finger and3 implications for "jumping-linker" dna recognition
13 d7znfa_
42.9
46
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H214 d1pvma3
41.1
44
Fold: Rubredoxin-likeSuperfamily: Hypothetical protein Ta0289 C-terminal domainFamily: Hypothetical protein Ta0289 C-terminal domain15 c1dgsB_
39.2
26
PDB header: ligaseChain: B: PDB Molecule: dna ligase;PDBTitle: crystal structure of nad+-dependent dna ligase from t.2 filiformis
16 c2elpA_
38.7
37
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 13th c2h2 zinc finger of human2 zinc finger protein 406
17 d1vq0a2
38.3
18
Fold: HSP33 redox switch-likeSuperfamily: HSP33 redox switch-likeFamily: HSP33 redox switch-like18 c2l8eA_
37.6
40
PDB header: dna binding proteinChain: A: PDB Molecule: polyhomeotic-like protein 1;PDBTitle: solution nmr structure of fcs domain of human polyhomeotic homolog 12 (hph1)
19 c1rikA_
37.4
27
PDB header: de novo proteinChain: A: PDB Molecule: e6apc1 peptide;PDBTitle: e6-binding zinc finger (e6apc1)
20 c5znfA_
37.3
57
PDB header: zinc finger dna binding domainChain: A: PDB Molecule: zinc finger;PDBTitle: alternating zinc fingers in the human male associated2 protein zfy: 2d nmr structure of an even finger and3 implications for "jumping-linker" dna recognition
21 d1xjha_
not modelled
35.6
29
Fold: HSP33 redox switch-likeSuperfamily: HSP33 redox switch-likeFamily: HSP33 redox switch-like22 c2jvmA_
not modelled
35.2
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of rhodobacter sphaeroides protein2 rhos4_26430. northeast structural genomics consortium3 target rhr95
23 c1vzyA_
not modelled
34.9
12
PDB header: chaperoneChain: A: PDB Molecule: 33 kda chaperonin;PDBTitle: crystal structure of the bacillus subtilis hsp33
24 d1vzya2
not modelled
34.1
12
Fold: HSP33 redox switch-likeSuperfamily: HSP33 redox switch-likeFamily: HSP33 redox switch-like25 c3siqF_
not modelled
33.4
17
PDB header: ligaseChain: F: PDB Molecule: apoptosis 1 inhibitor;PDBTitle: crystal structure of autoinhibited diap1-bir1 domain
26 c2jrrA_
not modelled
32.7
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of q5lls5 from silicibacter2 pomeroyi. northeast structural genomics consortium target3 sir90
27 c1vq0A_
not modelled
32.1
17
PDB header: chaperoneChain: A: PDB Molecule: 33 kda chaperonin;PDBTitle: crystal structure of 33 kda chaperonin (heat shock protein 33 homolog)2 (hsp33) (tm1394) from thermotoga maritima at 2.20 a resolution
28 c1klsA_
not modelled
29.8
57
PDB header: transcriptionChain: A: PDB Molecule: zinc finger y-chromosomal protein;PDBTitle: nmr structure of the zfy-6t[y10l] zinc finger
29 c1klrA_
not modelled
29.6
57
PDB header: transcriptionChain: A: PDB Molecule: zinc finger y-chromosomal protein;PDBTitle: nmr structure of the zfy-6t[y10f] zinc finger
30 d1klra_
not modelled
29.6
57
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H231 d1njqa_
not modelled
28.0
29
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Plant C2H2 finger (QALGGH zinc finger)32 c2elvA_
not modelled
27.5
22
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 6th c2h2 zinc finger of human2 zinc finger protein 406
33 d2cota1
not modelled
27.0
42
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H234 d1a1ia2
not modelled
26.9
24
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H235 d1llmc2
not modelled
26.6
18
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H236 c2eltA_
not modelled
25.3
19
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 3rd c2h2 zinc finger of human2 zinc finger protein 406
37 d2ct1a2
not modelled
25.2
20
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H238 c1arfA_
not modelled
25.1
21
PDB header: transcription regulationChain: A: PDB Molecule: yeast transcription factor adr1;PDBTitle: structures of dna-binding mutant zinc finger domains:2 implications for dna binding
39 c2jz8A_
not modelled
24.2
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein bh09830;PDBTitle: solution nmr structure of bh09830 from bartonella henselae2 modeled with one zn+2 bound. northeast structural genomics3 consortium target bnr55
40 d1ncsa_
not modelled
24.1
28
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H241 c1meyG_
not modelled
23.9
38
PDB header: transferase/dnaChain: G: PDB Molecule: consensus zinc finger;PDBTitle: crystal structure of a designed zinc finger protein bound2 to dna
42 c2eprA_
not modelled
22.8
22
PDB header: transcriptionChain: A: PDB Molecule: poz-, at hook-, and zinc finger-containingPDBTitle: solution structure of the secound zinc finger domain of2 zinc finger protein 278
43 c3t6pA_
not modelled
22.6
25
PDB header: apoptosisChain: A: PDB Molecule: baculoviral iap repeat-containing protein 2;PDBTitle: iap antagonist-induced conformational change in ciap1 promotes e32 ligase activation via dimerization
44 d1u85a1
not modelled
22.3
36
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H245 c2elqA_
not modelled
21.7
25
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 14th c2h2 zinc finger of human2 zinc finger protein 406
46 c2drpD_
not modelled
21.4
20
PDB header: transcription/dnaChain: D: PDB Molecule: protein (tramtrack dna-binding domain);PDBTitle: the crystal structure of a two zinc-finger peptide reveals2 an extension to the rules for zinc-finger/dna recognition
47 d1m4ma_
not modelled
21.2
22
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat48 d1tfqa_
not modelled
21.1
19
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat49 c2el5A_
not modelled
21.0
33
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 268;PDBTitle: solution structure of the 18th zf-c2h2 domain from human2 zinc finger protein 268
50 c3m0aD_
not modelled
20.5
19
PDB header: signaling proteinChain: D: PDB Molecule: baculoviral iap repeat-containing protein 3;PDBTitle: crystal structure of traf2:ciap2 complex
51 d1i3oe_
not modelled
20.3
10
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat52 d2raxa1
not modelled
20.1
19
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat53 d1xb0a_
not modelled
20.1
9
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat54 d1srka_
not modelled
20.0
18
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H255 d2dkta1
not modelled
20.0
23
Fold: CHY zinc finger-likeSuperfamily: CHY zinc finger-likeFamily: CHY zinc finger56 d2vsla1
not modelled
19.7
12
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat57 d1a1ia3
not modelled
19.3
18
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H258 d1se0a_
not modelled
19.3
19
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat59 d1q4qa_
not modelled
19.0
13
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat60 d1m4ka2
not modelled
18.9
12
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains61 d2i3ha1
not modelled
18.8
14
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat62 d1dgsa1
not modelled
18.8
56
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: NAD+-dependent DNA ligase, domain 363 d1tf3a2
not modelled
18.4
43
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H264 d3d9ta1
not modelled
18.4
29
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat65 d2epsa1
not modelled
18.1
42
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H266 d1xb0b_
not modelled
18.1
14
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat67 d2epra1
not modelled
18.1
36
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H268 c1v9pB_
not modelled
17.9
24
PDB header: ligaseChain: B: PDB Molecule: dna ligase;PDBTitle: crystal structure of nad+-dependent dna ligase
69 d1x6ha2
not modelled
17.4
26
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H270 c2poiA_
not modelled
16.9
13
PDB header: signaling protein/apoptosisChain: A: PDB Molecule: baculoviral iap repeat-containing protein 4;PDBTitle: crystal structure of xiap bir1 domain (i222 form)
71 c2elsA_
not modelled
16.9
28
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 2nd c2h2 zinc finger of human2 zinc finger protein 406
72 d1g73d_
not modelled
16.8
19
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat73 d1jd5a_
not modelled
16.6
14
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat74 c1va3A_
not modelled
16.5
29
PDB header: transcriptionChain: A: PDB Molecule: transcription factor sp1;PDBTitle: solution structure of transcription factor sp1 dna binding2 domain (zinc finger 3)
75 d2vrda1
not modelled
16.5
27
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: HkH motif-containing C2H2 finger76 d1oxna_
not modelled
16.4
14
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat77 d1x6ea2
not modelled
16.2
50
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H278 d1vd4a_
not modelled
16.1
14
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain79 c1rimA_
not modelled
16.0
27
PDB header: de novo proteinChain: A: PDB Molecule: e6apc2 peptide;PDBTitle: e6-binding zinc finger (e6apc2)
80 c2yu5A_
not modelled
15.7
31
PDB header: rna binding proteinChain: A: PDB Molecule: zinc finger protein 473;PDBTitle: solution structure of the zf-c2h2 domain (669-699aa) in2 zinc finger protein 473
81 c2eoxA_
not modelled
15.7
21
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 473;PDBTitle: solution structure of the c2h2 type zinc finger (region 315-2 345) of human zinc finger protein 473
82 d1xf7a_
not modelled
15.7
45
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H283 c1xf7A_
not modelled
15.7
45
PDB header: transcriptionChain: A: PDB Molecule: wilms' tumor protein;PDBTitle: high resolution nmr structure of the wilms' tumor2 suppressor protein (wt1) finger 3
84 d2qfaa1
not modelled
15.6
19
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat85 c2epcA_
not modelled
15.6
21
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 32;PDBTitle: solution structure of zinc finger domain 7 in zinc finger2 protein 32
86 d1nkra2
not modelled
15.6
12
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains87 c2eifA_
not modelled
15.3
13
PDB header: gene regulationChain: A: PDB Molecule: protein (eukaryotic translation initiation factor 5a);PDBTitle: eukaryotic translation initiation factor 5a from methanococcus2 jannaschii
88 d2drpa1
not modelled
15.3
20
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H289 c2epuA_
not modelled
15.1
33
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 32;PDBTitle: solution structure of the secound c2h2 type zinc finger2 domain of zinc finger protein 32
90 c1oy7C_
not modelled
14.9
14
PDB header: apoptosis/peptideChain: C: PDB Molecule: baculoviral iap repeat-containing protein 7;PDBTitle: structure and function analysis of peptide antagonists of melanoma2 inhibitor of apoptosis (ml-iap)
91 c2en3A_
not modelled
14.6
25
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 95 homolog;PDBTitle: solution structure of the c2h2 type zinc finger (region 796-2 828) of human zinc finger protein 95 homolog
92 c2vm5A_
not modelled
14.5
14
PDB header: apoptosisChain: A: PDB Molecule: baculoviral iap repeat-containing protein 1;PDBTitle: human bir2 domain of baculoviral inhibitor of apoptosis2 repeat-containing 1 (birc1)
93 d1ueba1
not modelled
14.2
10
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: eIF5a N-terminal domain-like94 c3m1cA_
not modelled
14.0
22
PDB header: viral proteinChain: A: PDB Molecule: envelope glycoprotein h;PDBTitle: crystal structure of the conserved herpesvirus fusion regulator2 complex gh-gl
95 d1ubdc3
not modelled
14.0
42
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H296 d1sp1a_
not modelled
13.9
29
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H297 c1sp1A_
not modelled
13.9
29
PDB header: zinc fingerChain: A: PDB Molecule: sp1f3;PDBTitle: nmr structure of a zinc finger domain from transcription2 factor sp1f3, minimized average structure
98 d2dl2a1
not modelled
13.2
12
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains99 c2en8A_
not modelled
13.1
20
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 224;PDBTitle: solution structure of the c2h2 type zinc finger (region 171-2 203) of human zinc finger protein 224