| 1 |
|
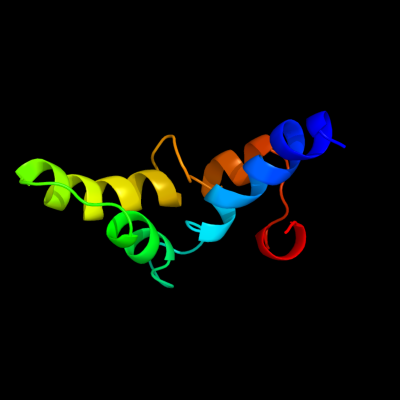
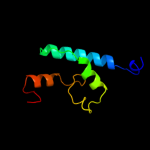
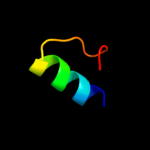
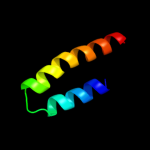
PDB 1z9h chain A domain 1
Region: 67 - 143
Aligned: 69
Modelled: 77
Confidence: 37.0%
Identity: 23%
Fold: GST C-terminal domain-like
Superfamily: GST C-terminal domain-like
Family: Glutathione S-transferase (GST), C-terminal domain
Phyre2
| 2 |
|
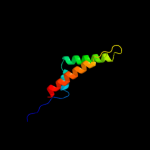
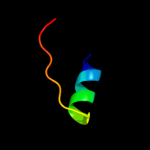
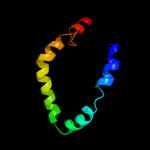
PDB 3n8u chain B
Region: 129 - 250
Aligned: 117
Modelled: 122
Confidence: 34.8%
Identity: 15%
PDB header:hydrolase
Chain: B: PDB Molecule:imelysin peptidase;
PDBTitle: crystal structure of an imelysin peptidase (bacova_03801) from2 bacteroides ovatus at 1.44 a resolution
Phyre2
| 3 |
|
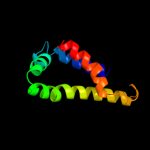
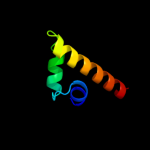
PDB 1tq4 chain A
Region: 165 - 249
Aligned: 84
Modelled: 85
Confidence: 31.1%
Identity: 7%
Fold: P-loop containing nucleoside triphosphate hydrolases
Superfamily: P-loop containing nucleoside triphosphate hydrolases
Family: G proteins
Phyre2
| 4 |
|
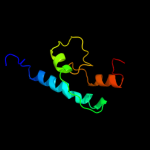
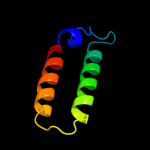
PDB 1qs0 chain A
Region: 176 - 254
Aligned: 79
Modelled: 79
Confidence: 29.5%
Identity: 13%
Fold: Thiamin diphosphate-binding fold (THDP-binding)
Superfamily: Thiamin diphosphate-binding fold (THDP-binding)
Family: Branched-chain alpha-keto acid dehydrogenase PP module
Phyre2
| 5 |
|
PDB 1wpg chain A domain 4
Region: 57 - 124
Aligned: 68
Modelled: 68
Confidence: 22.5%
Identity: 13%
Fold: Calcium ATPase, transmembrane domain M
Superfamily: Calcium ATPase, transmembrane domain M
Family: Calcium ATPase, transmembrane domain M
Phyre2
| 6 |
|
PDB 2id3 chain A domain 2
Region: 146 - 239
Aligned: 93
Modelled: 94
Confidence: 17.7%
Identity: 12%
Fold: Tetracyclin repressor-like, C-terminal domain
Superfamily: Tetracyclin repressor-like, C-terminal domain
Family: Tetracyclin repressor-like, C-terminal domain
Phyre2
| 7 |
|
PDB 2fq4 chain A domain 2
Region: 149 - 239
Aligned: 87
Modelled: 91
Confidence: 11.2%
Identity: 13%
Fold: Tetracyclin repressor-like, C-terminal domain
Superfamily: Tetracyclin repressor-like, C-terminal domain
Family: Tetracyclin repressor-like, C-terminal domain
Phyre2
| 8 |
|
PDB 2bfd chain A domain 1
Region: 176 - 254
Aligned: 76
Modelled: 79
Confidence: 10.2%
Identity: 17%
Fold: Thiamin diphosphate-binding fold (THDP-binding)
Superfamily: Thiamin diphosphate-binding fold (THDP-binding)
Family: Branched-chain alpha-keto acid dehydrogenase PP module
Phyre2
| 9 |
|
PDB 3ku7 chain B
Region: 327 - 348
Aligned: 22
Modelled: 22
Confidence: 9.6%
Identity: 14%
PDB header:cell cycle
Chain: B: PDB Molecule:cell division topological specificity factor;
PDBTitle: crystal structure of helicobacter pylori mine, a cell division2 topological specificity factor
Phyre2
| 10 |
|
PDB 1mhs chain A
Region: 10 - 131
Aligned: 115
Modelled: 122
Confidence: 9.4%
Identity: 11%
PDB header:membrane protein, proton transport
Chain: A: PDB Molecule:plasma membrane atpase;
PDBTitle: model of neurospora crassa proton atpase
Phyre2
| 11 |
|
PDB 2voy chain B
Region: 57 - 94
Aligned: 38
Modelled: 38
Confidence: 8.6%
Identity: 16%
PDB header:hydrolase
Chain: B: PDB Molecule:sarcoplasmic/endoplasmic reticulum calcium
PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
Phyre2
| 12 |
|
PDB 1qhb chain A
Region: 312 - 342
Aligned: 31
Modelled: 31
Confidence: 8.4%
Identity: 26%
Fold: Acid phosphatase/Vanadium-dependent haloperoxidase
Superfamily: Acid phosphatase/Vanadium-dependent haloperoxidase
Family: Haloperoxidase (bromoperoxidase)
Phyre2
| 13 |
|
PDB 3trb chain A
Region: 235 - 253
Aligned: 19
Modelled: 19
Confidence: 6.7%
Identity: 16%
PDB header:dna binding protein
Chain: A: PDB Molecule:virulence-associated protein i;
PDBTitle: structure of an addiction module antidote protein of a higa (higa)2 family from coxiella burnetii
Phyre2
| 14 |
|
PDB 2xyk chain B
Region: 159 - 211
Aligned: 53
Modelled: 53
Confidence: 6.2%
Identity: 6%
PDB header:oxygen storage/transport
Chain: B: PDB Molecule:2-on-2 hemoglobin;
PDBTitle: group ii 2-on-2 hemoglobin from the plant pathogen2 agrobacterium tumefaciens
Phyre2
| 15 |
|
PDB 2eby chain A
Region: 235 - 253
Aligned: 19
Modelled: 19
Confidence: 6.0%
Identity: 11%
PDB header:transcription
Chain: A: PDB Molecule:putative hth-type transcriptional regulator ybaq;
PDBTitle: crystal structure of a hypothetical protein from e. coli
Phyre2
| 16 |
|
PDB 2ict chain A domain 1
Region: 235 - 253
Aligned: 19
Modelled: 19
Confidence: 6.0%
Identity: 5%
Fold: lambda repressor-like DNA-binding domains
Superfamily: lambda repressor-like DNA-binding domains
Family: SinR domain-like
Phyre2
| 17 |
|
PDB 2ap1 chain A domain 1
Region: 171 - 219
Aligned: 44
Modelled: 49
Confidence: 5.9%
Identity: 14%
Fold: Ribonuclease H-like motif
Superfamily: Actin-like ATPase domain
Family: ROK
Phyre2
| 18 |
|
PDB 3few chain X
Region: 67 - 120
Aligned: 54
Modelled: 54
Confidence: 5.8%
Identity: 15%
PDB header:immune system
Chain: X: PDB Molecule:colicin s4;
PDBTitle: structure and function of colicin s4, a colicin with a2 duplicated receptor binding domain
Phyre2
| 19 |
|
PDB 2rpa chain A
Region: 152 - 189
Aligned: 38
Modelled: 38
Confidence: 5.3%
Identity: 11%
PDB header:hydrolase
Chain: A: PDB Molecule:katanin p60 atpase-containing subunit a1;
PDBTitle: the solution structure of n-terminal domain of microtubule severing2 enzyme
Phyre2
| 20 |
|
PDB 1o8t chain A
Region: 171 - 233
Aligned: 59
Modelled: 63
Confidence: 5.2%
Identity: 19%
PDB header:lipid transport
Chain: A: PDB Molecule:apolipoprotein c-ii;
PDBTitle: global structure and dynamics of human apolipoprotein cii2 in complex with micelles: evidence for increased mobility3 of the helix involvved in the activation of lipoprotein4 lipase.
Phyre2
| 21 |
|
| 22 |
|