| 1 | c1rg9D_ |
|
 |
100.0 |
100 |
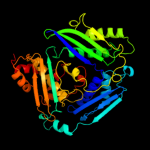
PDB header:transferase
Chain: D: PDB Molecule:s-adenosylmethionine synthetase;
PDBTitle: s-adenosylmethionine synthetase complexed with sam and ppnp
|
| 2 | c2obvA_ |
|
 |
100.0 |
57 |
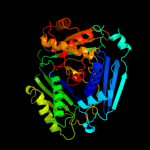
PDB header:transferase
Chain: A: PDB Molecule:s-adenosylmethionine synthetase isoform type-1;
PDBTitle: crystal structure of the human s-adenosylmethionine synthetase 1 in2 complex with the product
|
| 3 | c3so4C_ |
|
 |
100.0 |
52 |
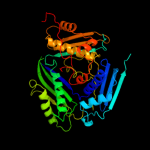
PDB header:transferase
Chain: C: PDB Molecule:methionine-adenosyltransferase;
PDBTitle: methionine-adenosyltransferase from entamoeba histolytica
|
| 4 | c3imlB_ |
|
 |
100.0 |
72 |
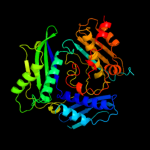
PDB header:transferase
Chain: B: PDB Molecule:s-adenosylmethionine synthetase;
PDBTitle: crystal structure of s-adenosylmethionine synthetase from burkholderia2 pseudomallei
|
| 5 | c3rv2B_ |
|
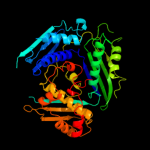 |
100.0 |
62 |
PDB header:transferase
Chain: B: PDB Molecule:s-adenosylmethionine synthase;
PDBTitle: crystal structure of s-adenosylmethionine synthetase from2 mycobacterium marinum
|
| 6 | d1mxaa3 |
|
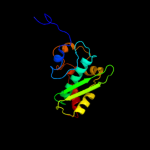 |
100.0 |
100 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 7 | d2p02a3 |
|
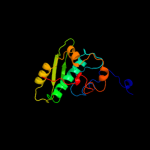 |
100.0 |
62 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 8 | d1qm4a3 |
|
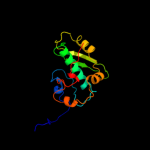 |
100.0 |
64 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 9 | d1mxaa2 |
|
 |
100.0 |
100 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 10 | d2p02a2 |
|
 |
100.0 |
48 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 11 | d1qm4a2 |
|
 |
100.0 |
49 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 12 | d2p02a1 |
|
 |
100.0 |
61 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 13 | d1mxaa1 |
|
 |
100.0 |
100 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 14 | d1qm4a1 |
|
 |
100.0 |
62 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 15 | c3k8zD_ |
|
 |
82.9 |
13 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:nad-specific glutamate dehydrogenase;
PDBTitle: crystal structure of gudb1 a decryptified secondary glutamate2 dehydrogenase from b. subtilis
|
| 16 | d2dt5a1 |
|
 |
82.1 |
28 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Transcriptional repressor Rex, N-terminal domain |
| 17 | c2bmaA_ |
|
 |
56.4 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutamate dehydrogenase (nadp+);
PDBTitle: the crystal structure of plasmodium falciparum glutamate2 dehydrogenase, a putative target for novel antimalarial3 drugs
|
| 18 | d1w9ha1 |
|
 |
54.5 |
23 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:PIWI domain |
| 19 | c3lvtA_ |
|
 |
51.4 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:glycosyl hydrolase, family 38;
PDBTitle: the crystal structure of a protein in the glycosyl hydrolase2 family 38 from enterococcus faecalis to 2.55a
|
| 20 | c2inrA_ |
|
 |
48.9 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:dna topoisomerase 4 subunit a;
PDBTitle: crystal structure of a 59 kda fragment of topoisomerase iv subunit a2 (grla) from staphylococcus aureus
|
| 21 | c2w42A_ |
|
not modelled |
40.4 |
24 |
PDB header:protein/dna complex
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: the structure of a piwi protein from archaeoglobus fulgidus2 complexed with a 16nt dna duplex.
|
| 22 | d1u04a2 |
|
not modelled |
39.0 |
31 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:PIWI domain |
| 23 | d1euza2 |
|
not modelled |
36.1 |
18 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
| 24 | c3ketA_ |
|
not modelled |
34.9 |
17 |
PDB header:transcription/dna
Chain: A: PDB Molecule:redox-sensing transcriptional repressor rex;
PDBTitle: crystal structure of a rex-family transcriptional regulatory protein2 from streptococcus agalactiae bound to a palindromic operator
|
| 25 | d1kjna_ |
|
not modelled |
34.4 |
29 |
Fold:Hypothetical protein MTH777 (MT0777)
Superfamily:Hypothetical protein MTH777 (MT0777)
Family:Hypothetical protein MTH777 (MT0777) |
| 26 | c3aoeC_ |
|
not modelled |
31.1 |
20 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glutamate dehydrogenase;
PDBTitle: crystal structure of hetero-hexameric glutamate dehydrogenase from2 thermus thermophilus (leu bound form)
|
| 27 | d2fiqa1 |
|
not modelled |
28.4 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:GatZ-like |
| 28 | c2dt5A_ |
|
not modelled |
27.9 |
28 |
PDB header:dna binding protein
Chain: A: PDB Molecule:at-rich dna-binding protein;
PDBTitle: crystal structure of ttha1657 (at-rich dna-binding protein) from2 thermus thermophilus hb8
|
| 29 | d1to0a_ |
|
not modelled |
24.0 |
60 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YbeA-like |
| 30 | d2fm8a1 |
|
not modelled |
23.6 |
22 |
Fold:Secretion chaperone-like
Superfamily:Type III secretory system chaperone-like
Family:Type III secretory system chaperone |
| 31 | c2vofA_ |
|
not modelled |
22.7 |
8 |
PDB header:apoptosis
Chain: A: PDB Molecule:bcl-2-related protein a1;
PDBTitle: structure of mouse a1 bound to the puma bh3-domain
|
| 32 | d1vh0a_ |
|
not modelled |
22.7 |
75 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YbeA-like |
| 33 | c1hrdA_ |
|
not modelled |
22.2 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutamate dehydrogenase;
PDBTitle: glutamate dehydrogenase
|
| 34 | d1ns5a_ |
|
not modelled |
22.0 |
50 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YbeA-like |
| 35 | d1o6da_ |
|
not modelled |
21.3 |
63 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YbeA-like |
| 36 | d1b26a2 |
|
not modelled |
20.9 |
23 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
| 37 | c2z51A_ |
|
not modelled |
20.1 |
20 |
PDB header:metal transport
Chain: A: PDB Molecule:nifu-like protein 2, chloroplast;
PDBTitle: crystal structure of arabidopsis cnfu involved in iron-2 sulfur cluster biosynthesis
|
| 38 | d1vkha_ |
|
not modelled |
19.8 |
19 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Putative serine hydrolase Ydr428c |
| 39 | d1azpa_ |
|
not modelled |
17.4 |
40 |
Fold:SH3-like barrel
Superfamily:Chromo domain-like
Family:"Histone-like" proteins from archaea |
| 40 | c1u04A_ |
|
not modelled |
17.3 |
46 |
PDB header:hydrolase/gene regulation
Chain: A: PDB Molecule:hypothetical protein pf0537;
PDBTitle: crystal structure of full length argonaute from pyrococcus furiosus
|
| 41 | c1v57A_ |
|
not modelled |
16.5 |
26 |
PDB header:isomerase
Chain: A: PDB Molecule:thiol:disulfide interchange protein dsbg;
PDBTitle: crystal structure of the disulfide bond isomerase dsbg
|
| 42 | c2jvfA_ |
|
not modelled |
15.4 |
9 |
PDB header:de novo protein
Chain: A: PDB Molecule:de novo protein m7;
PDBTitle: solution structure of m7, a computationally-designed2 artificial protein
|
| 43 | d1q33a_ |
|
not modelled |
15.3 |
60 |
Fold:Nudix
Superfamily:Nudix
Family:MutT-like |
| 44 | c2bh7A_ |
|
not modelled |
14.4 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-acetylmuramoyl-l-alanine amidase;
PDBTitle: crystal structure of a semet derivative of amid at 2.22 angstroms
|
| 45 | d1gtma2 |
|
not modelled |
14.1 |
13 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
| 46 | c3lwaA_ |
|
not modelled |
13.9 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:secreted thiol-disulfide isomerase;
PDBTitle: the crystal structure of a secreted thiol-disulfide2 isomerase from corynebacterium glutamicum to 1.75a
|
| 47 | c3h6nA_ |
|
not modelled |
13.7 |
14 |
PDB header:signaling protein
Chain: A: PDB Molecule:plexin-d1;
PDBTitle: crystal structure of the ubiquitin-like domain of plexin d1
|
| 48 | d2f9wa2 |
|
not modelled |
13.5 |
50 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:CoaX-like |
| 49 | c2gjhA_ |
|
not modelled |
13.1 |
18 |
PDB header:de novo protein
Chain: A: PDB Molecule:designed protein;
PDBTitle: nmr structure of cfr (c-terminal fragment of2 computationally designed novel-topology protein top7)
|
| 50 | c2fmmE_ |
|
not modelled |
13.0 |
21 |
PDB header:transcription
Chain: E: PDB Molecule:protein emsy;
PDBTitle: crystal structure of emsy-hp1 complex
|
| 51 | d1lfda_ |
|
not modelled |
12.9 |
17 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ras-binding domain, RBD |
| 52 | d2fmme1 |
|
not modelled |
12.9 |
21 |
Fold:ENT-like
Superfamily:ENT-like
Family:Emsy N terminal (ENT) domain-like |
| 53 | d1vhfa_ |
|
not modelled |
12.8 |
11 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Divalent ion tolerance proteins CutA (CutA1) |
| 54 | d1t0ga_ |
|
not modelled |
12.7 |
39 |
Fold:Cytochrome b5-like heme/steroid binding domain
Superfamily:Cytochrome b5-like heme/steroid binding domain
Family:Steroid-binding domain |
| 55 | c1wnkA_ |
|
not modelled |
12.4 |
50 |
PDB header:transcription
Chain: A: PDB Molecule:fibroin-modulator-binding-protein-1;
PDBTitle: nmr structure of fmbp-1 tandem repeat 3 in 30%(v/v) tfe2 solution
|
| 56 | d1uz3a1 |
|
not modelled |
12.3 |
21 |
Fold:ENT-like
Superfamily:ENT-like
Family:Emsy N terminal (ENT) domain-like |
| 57 | d1knga_ |
|
not modelled |
12.2 |
8 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 58 | d1ztpa1 |
|
not modelled |
12.0 |
16 |
Fold:eIF4e-like
Superfamily:eIF4e-like
Family:BLES03-like |
| 59 | c2jkzB_ |
|
not modelled |
12.0 |
44 |
PDB header:transferase
Chain: B: PDB Molecule:hypoxanthine-guanine phosphoribosyltransferase;
PDBTitle: saccharomyces cerevisiae hypoxanthine-guanine2 phosphoribosyltransferase in complex with gmp (guanosine 5'3 -monophosphate) (orthorhombic crystal form)
|
| 60 | d1p1la_ |
|
not modelled |
11.5 |
32 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Divalent ion tolerance proteins CutA (CutA1) |
| 61 | d1yvua2 |
|
not modelled |
11.5 |
18 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:PIWI domain |
| 62 | c2tmgD_ |
|
not modelled |
11.4 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:protein (glutamate dehydrogenase);
PDBTitle: thermotoga maritima glutamate dehydrogenase mutant s128r,2 t158e, n117r, s160e
|
| 63 | d2bu3a1 |
|
not modelled |
11.2 |
26 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Phytochelatin synthase |
| 64 | c2zfhA_ |
|
not modelled |
11.1 |
26 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:cuta;
PDBTitle: crystal structure of putative cuta1 from homo sapiens at 2.05a2 resolution
|
| 65 | c3kh0A_ |
|
not modelled |
11.1 |
19 |
PDB header:signaling protein
Chain: A: PDB Molecule:ral guanine nucleotide dissociation stimulator;
PDBTitle: crystal structure of the ras-association (ra) domain of2 ralgds
|
| 66 | c2zomC_ |
|
not modelled |
11.0 |
26 |
PDB header:unknown function
Chain: C: PDB Molecule:protein cuta, chloroplast, putative, expressed;
PDBTitle: crystal structure of cuta1 from oryza sativa
|
| 67 | d1ah4a_ |
|
not modelled |
11.0 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 68 | c1qysA_ |
|
not modelled |
10.6 |
25 |
PDB header:de novo protein
Chain: A: PDB Molecule:top7;
PDBTitle: crystal structure of top7: a computationally designed2 protein with a novel fold
|
| 69 | c2wl2B_ |
|
not modelled |
10.6 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:dna gyrase subunit a;
PDBTitle: crystal structure of n-terminal domain of gyra with the2 antibiotic simocyclinone d8
|
| 70 | d2bgxa2 |
|
not modelled |
10.5 |
26 |
Fold:N-acetylmuramoyl-L-alanine amidase-like
Superfamily:N-acetylmuramoyl-L-alanine amidase-like
Family:N-acetylmuramoyl-L-alanine amidase-like |
| 71 | d1naqa_ |
|
not modelled |
10.1 |
26 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Divalent ion tolerance proteins CutA (CutA1) |
| 72 | d1bf4a_ |
|
not modelled |
10.0 |
48 |
Fold:SH3-like barrel
Superfamily:Chromo domain-like
Family:"Histone-like" proteins from archaea |
| 73 | d1rlfa_ |
|
not modelled |
9.5 |
24 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ras-binding domain, RBD |
| 74 | d1nzaa_ |
|
not modelled |
9.4 |
42 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Divalent ion tolerance proteins CutA (CutA1) |
| 75 | c2btwA_ |
|
not modelled |
9.2 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:alr0975 protein;
PDBTitle: crystal structure of alr0975
|
| 76 | c3ag7A_ |
|
not modelled |
9.0 |
35 |
PDB header:plant protein
Chain: A: PDB Molecule:putative uncharacterized protein f9e10.5;
PDBTitle: an auxilin-like j-domain containing protein, jac1 j-domain
|
| 77 | d2rgfa_ |
|
not modelled |
9.0 |
19 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ras-binding domain, RBD |
| 78 | c2nuhA_ |
|
not modelled |
8.9 |
21 |
PDB header:unknown function
Chain: A: PDB Molecule:periplasmic divalent cation tolerance protein;
PDBTitle: crystal structure of cuta from the phytopathgen bacterium xylella2 fastidiosa
|
| 79 | c3gdeA_ |
|
not modelled |
8.9 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:dna ligase;
PDBTitle: the closed conformation of atp-dependent dna ligase from2 archaeoglobus fulgidus
|
| 80 | d3c9fa1 |
|
not modelled |
8.8 |
16 |
Fold:5'-nucleotidase (syn. UDP-sugar hydrolase), C-terminal domain
Superfamily:5'-nucleotidase (syn. UDP-sugar hydrolase), C-terminal domain
Family:5'-nucleotidase (syn. UDP-sugar hydrolase), C-terminal domain |
| 81 | c2kucA_ |
|
not modelled |
8.7 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:putative disulphide-isomerase;
PDBTitle: solution structure of a putative disulphide-isomerase from bacteroides2 thetaiotaomicron
|
| 82 | d1osce_ |
|
not modelled |
8.7 |
26 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Divalent ion tolerance proteins CutA (CutA1) |
| 83 | c3b6nA_ |
|
not modelled |
8.7 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:2-c-methyl-d-erythritol 2,4-cyclodiphosphate
PDBTitle: crystal structure of 2c-methyl-d-erythritol 2,4-2 cyclodiphosphate synthase pv003920 from plasmodium vivax
|
| 84 | c3lorB_ |
|
not modelled |
8.6 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:thiol-disulfide isomerase and thioredoxins;
PDBTitle: the crystal structure of a thiol-disulfide isomerase from2 corynebacterium glutamicum to 2.2a
|
| 85 | d1j03a_ |
|
not modelled |
8.4 |
39 |
Fold:Cytochrome b5-like heme/steroid binding domain
Superfamily:Cytochrome b5-like heme/steroid binding domain
Family:Steroid-binding domain |
| 86 | d1hkoa_ |
|
not modelled |
8.4 |
36 |
Fold:Cytochrome b5-like heme/steroid binding domain
Superfamily:Cytochrome b5-like heme/steroid binding domain
Family:Cytochrome b5 |
| 87 | d2zfha1 |
|
not modelled |
8.4 |
26 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Divalent ion tolerance proteins CutA (CutA1) |
| 88 | c1xk8A_ |
|
not modelled |
8.4 |
26 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:divalent cation tolerant protein cuta;
PDBTitle: divalent cation tolerant protein cuta from homo sapiens2 o60888
|
| 89 | d1ukua_ |
|
not modelled |
8.4 |
21 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Divalent ion tolerance proteins CutA (CutA1) |
| 90 | c2wnyB_ |
|
not modelled |
8.2 |
17 |
PDB header:unknown function
Chain: B: PDB Molecule:conserved protein mth689;
PDBTitle: structure of mth689, a duf54 protein from methanothermobacter2 thermautotrophicus
|
| 91 | d1c9ka_ |
|
not modelled |
8.2 |
86 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 92 | c3d4oA_ |
|
not modelled |
8.1 |
44 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit a;
PDBTitle: crystal structure of dipicolinate synthase subunit a (np_243269.1)2 from bacillus halodurans at 2.10 a resolution
|
| 93 | d1mhyd_ |
|
not modelled |
8.0 |
35 |
Fold:Ferritin-like
Superfamily:Ferritin-like
Family:Ribonucleotide reductase-like |
| 94 | d1vjpa1 |
|
not modelled |
7.9 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 95 | c2hw2A_ |
|
not modelled |
7.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:rifampin adp-ribosyl transferase;
PDBTitle: crystal structure of rifampin adp-ribosyl transferase in2 complex with rifampin
|
| 96 | c3s5pA_ |
|
not modelled |
7.9 |
22 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: crystal structure of ribose-5-phosphate isomerase b rpib from giardia2 lamblia
|
| 97 | c3ga2A_ |
|
not modelled |
7.8 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:endonuclease v;
PDBTitle: crystal structure of the endonuclease_v (bsu36170) from2 bacillus subtilis, northeast structural genomics3 consortium target sr624
|
| 98 | d1kr4a_ |
|
not modelled |
7.8 |
11 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Divalent ion tolerance proteins CutA (CutA1) |
| 99 | d1mtyd_ |
|
not modelled |
7.7 |
41 |
Fold:Ferritin-like
Superfamily:Ferritin-like
Family:Ribonucleotide reductase-like |

