| 1 |
|
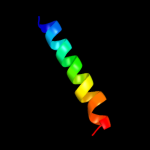
PDB 2fd6 chain U domain 2
Region: 80 - 95
Aligned: 16
Modelled: 16
Confidence: 16.1%
Identity: 25%
Fold: Snake toxin-like
Superfamily: Snake toxin-like
Family: Extracellular domain of cell surface receptors
Phyre2
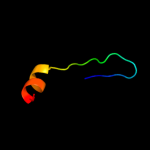
| 2 |
|
PDB 1v0d chain A
Region: 88 - 127
Aligned: 38
Modelled: 40
Confidence: 15.3%
Identity: 24%
Fold: His-Me finger endonucleases
Superfamily: His-Me finger endonucleases
Family: Caspase-activated DNase, CAD (DffB, DFF40)
Phyre2
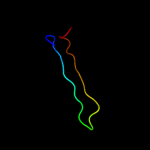
| 3 |
|
PDB 1v0d chain A
Region: 88 - 127
Aligned: 38
Modelled: 40
Confidence: 15.3%
Identity: 24%
PDB header:hydrolase
Chain: A: PDB Molecule:dna fragmentation factor 40 kda subunit;
PDBTitle: crystal structure of caspase-activated dnase (cad)
Phyre2
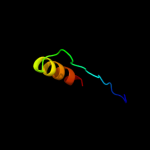
| 4 |
|
PDB 2vu9 chain A
Region: 12 - 47
Aligned: 33
Modelled: 36
Confidence: 12.3%
Identity: 27%
PDB header:hydrolase
Chain: A: PDB Molecule:botulinum neurotoxin a heavy chain;
PDBTitle: crystal structure of botulinum neurotoxin serotype a2 binding domain in complex with gt1b
Phyre2
| 5 |
|
PDB 3ts3 chain D
Region: 77 - 95
Aligned: 19
Modelled: 19
Confidence: 11.9%
Identity: 32%
PDB header:viral protein
Chain: D: PDB Molecule:capsid polyprotein;
PDBTitle: crystal structure of the projection domain of the turkey astrovirus2 capsid protein at 1.5 angstrom resolution
Phyre2
| 6 |
|
PDB 2lfc chain A
Region: 9 - 47
Aligned: 38
Modelled: 39
Confidence: 11.3%
Identity: 32%
PDB header:oxidoreductase
Chain: A: PDB Molecule:fumarate reductase, flavoprotein subunit;
PDBTitle: solution nmr structure of fumarate reductase flavoprotein subunit from2 lactobacillus plantarum, northeast structural genomics consortium3 target lpr145j
Phyre2
| 7 |
|
PDB 2w0c chain T
Region: 19 - 32
Aligned: 14
Modelled: 14
Confidence: 11.0%
Identity: 50%
PDB header:virus
Chain: T: PDB Molecule:protein p6;
PDBTitle: x-ray structure of the entire lipid-containing2 bacteriophage pm2
Phyre2
| 8 |
|
PDB 1p5d chain X domain 1
Region: 2 - 44
Aligned: 43
Modelled: 43
Confidence: 9.7%
Identity: 28%
Fold: Phosphoglucomutase, first 3 domains
Superfamily: Phosphoglucomutase, first 3 domains
Family: Phosphoglucomutase, first 3 domains
Phyre2
| 9 |
|
PDB 1hci chain A domain 1
Region: 14 - 37
Aligned: 24
Modelled: 24
Confidence: 8.8%
Identity: 25%
Fold: Spectrin repeat-like
Superfamily: Spectrin repeat
Family: Spectrin repeat
Phyre2
| 10 |
|
PDB 1g03 chain A
Region: 58 - 79
Aligned: 22
Modelled: 22
Confidence: 8.3%
Identity: 32%
Fold: Retrovirus capsid protein, N-terminal core domain
Superfamily: Retrovirus capsid protein, N-terminal core domain
Family: Retrovirus capsid protein, N-terminal core domain
Phyre2
| 11 |
|
PDB 2i9b chain F
Region: 80 - 109
Aligned: 30
Modelled: 30
Confidence: 8.0%
Identity: 20%
PDB header:hydrolase
Chain: F: PDB Molecule:urokinase plasminogen activator surface receptor;
PDBTitle: crystal structure of atf-urokinase receptor complex
Phyre2
| 12 |
|
PDB 1zy3 chain A domain 1
Region: 12 - 29
Aligned: 18
Modelled: 18
Confidence: 7.2%
Identity: 44%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 13 |
|
PDB 1o0l chain A
Region: 14 - 29
Aligned: 16
Modelled: 16
Confidence: 5.9%
Identity: 50%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 14 |
|
PDB 1pgl chain 2 domain 2
Region: 68 - 93
Aligned: 26
Modelled: 26
Confidence: 5.9%
Identity: 31%
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily: Positive stranded ssRNA viruses
Family: Comoviridae-like VP
Phyre2
| 15 |
|
PDB 1ny7 chain 2 domain 2
Region: 68 - 93
Aligned: 26
Modelled: 26
Confidence: 5.7%
Identity: 31%
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily: Positive stranded ssRNA viruses
Family: Comoviridae-like VP
Phyre2
| 16 |
|
PDB 1eij chain A
Region: 103 - 128
Aligned: 26
Modelled: 26
Confidence: 5.7%
Identity: 19%
Fold: RuvA C-terminal domain-like
Superfamily: Double-stranded DNA-binding domain
Family: Double-stranded DNA-binding domain
Phyre2
| 17 |
|
PDB 3jtp chain B
Region: 93 - 122
Aligned: 30
Modelled: 30
Confidence: 5.4%
Identity: 10%
PDB header:protein binding
Chain: B: PDB Molecule:adapter protein meca 1;
PDBTitle: crystal structure of the c-terminal domain of meca
Phyre2
| 18 |
|
PDB 2cru chain A domain 1
Region: 103 - 128
Aligned: 26
Modelled: 26
Confidence: 5.2%
Identity: 12%
Fold: RuvA C-terminal domain-like
Superfamily: Double-stranded DNA-binding domain
Family: Double-stranded DNA-binding domain
Phyre2