| 1 |
|
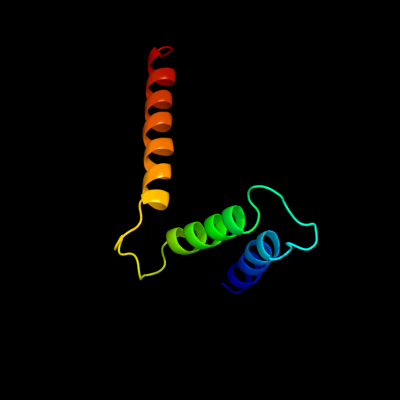
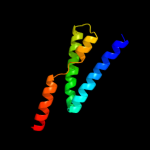
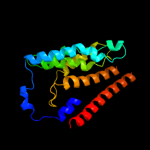
PDB 1s7b chain A
Region: 218 - 292
Aligned: 75
Modelled: 75
Confidence: 97.8%
Identity: 13%
Fold: Multidrug resistance efflux transporter EmrE
Superfamily: Multidrug resistance efflux transporter EmrE
Family: Multidrug resistance efflux transporter EmrE
Phyre2
| 2 |
|
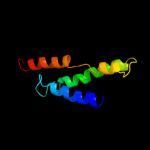
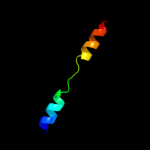
PDB 2i68 chain B
Region: 222 - 285
Aligned: 58
Modelled: 64
Confidence: 83.4%
Identity: 17%
PDB header:transport protein
Chain: B: PDB Molecule:protein emre;
PDBTitle: cryo-em based theoretical model structure of transmembrane2 domain of the multidrug-resistance antiporter from e. coli3 emre
Phyre2
| 3 |
|
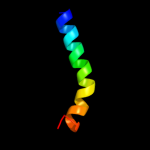
PDB 3aqp chain B
Region: 209 - 296
Aligned: 87
Modelled: 88
Confidence: 68.6%
Identity: 14%
PDB header:membrane protein
Chain: B: PDB Molecule:probable secdf protein-export membrane protein;
PDBTitle: crystal structure of secdf, a translocon-associated membrane protein,2 from thermus thrmophilus
Phyre2
| 4 |
|
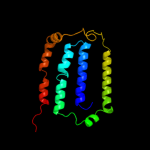
PDB 1iwg chain A domain 8
Region: 215 - 296
Aligned: 82
Modelled: 82
Confidence: 30.0%
Identity: 17%
Fold: Multidrug efflux transporter AcrB transmembrane domain
Superfamily: Multidrug efflux transporter AcrB transmembrane domain
Family: Multidrug efflux transporter AcrB transmembrane domain
Phyre2
| 5 |
|
PDB 2jp3 chain A
Region: 270 - 296
Aligned: 27
Modelled: 27
Confidence: 15.9%
Identity: 7%
PDB header:transcription
Chain: A: PDB Molecule:fxyd domain-containing ion transport regulator 4;
PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
Phyre2
| 6 |
|
PDB 3rko chain F
Region: 3 - 150
Aligned: 142
Modelled: 148
Confidence: 14.8%
Identity: 11%
PDB header:oxidoreductase
Chain: F: PDB Molecule:nadh-quinone oxidoreductase subunit j;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 7 |
|
PDB 3gia chain A
Region: 63 - 296
Aligned: 217
Modelled: 234
Confidence: 13.0%
Identity: 8%
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
Phyre2
| 8 |
|
PDB 3lrc chain C
Region: 183 - 294
Aligned: 112
Modelled: 112
Confidence: 8.0%
Identity: 10%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 9 |
|
PDB 2mlt chain B
Region: 271 - 295
Aligned: 25
Modelled: 25
Confidence: 7.1%
Identity: 4%
PDB header:toxin (hemolytic polypeptide)
Chain: B: PDB Molecule:melittin;
PDBTitle: melittin
Phyre2
| 10 |
|
PDB 2mlt chain A
Region: 271 - 295
Aligned: 25
Modelled: 25
Confidence: 7.1%
Identity: 4%
PDB header:toxin (hemolytic polypeptide)
Chain: A: PDB Molecule:melittin;
PDBTitle: melittin
Phyre2
| 11 |
|
PDB 3hd6 chain A
Region: 45 - 231
Aligned: 187
Modelled: 187
Confidence: 6.7%
Identity: 12%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 12 |
|
PDB 2hg5 chain D
Region: 269 - 296
Aligned: 28
Modelled: 28
Confidence: 6.0%
Identity: 11%
PDB header:membrane protein
Chain: D: PDB Molecule:kcsa channel;
PDBTitle: cs+ complex of a k channel with an amide to ester substitution in the2 selectivity filter
Phyre2
| 13 |
|
PDB 2knc chain B
Region: 263 - 296
Aligned: 34
Modelled: 34
Confidence: 5.1%
Identity: 15%
PDB header:cell adhesion
Chain: B: PDB Molecule:integrin beta-3;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2