| 1 |
|
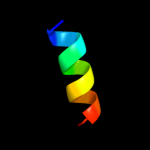
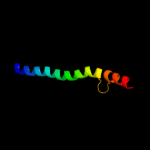
PDB 2p3x chain A
Region: 173 - 185
Aligned: 13
Modelled: 13
Confidence: 15.0%
Identity: 46%
PDB header:oxidoreductase
Chain: A: PDB Molecule:polyphenol oxidase, chloroplast;
PDBTitle: crystal structure of grenache (vitis vinifera) polyphenol2 oxidase
Phyre2
| 2 |
|
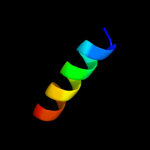
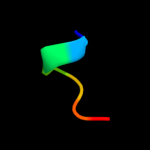
PDB 1bt3 chain A
Region: 173 - 185
Aligned: 13
Modelled: 13
Confidence: 14.1%
Identity: 38%
Fold: Di-copper centre-containing domain
Superfamily: Di-copper centre-containing domain
Family: Catechol oxidase
Phyre2
| 3 |
|
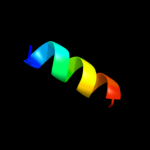
PDB 2knc chain A
Region: 152 - 205
Aligned: 39
Modelled: 39
Confidence: 12.6%
Identity: 10%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 4 |
|
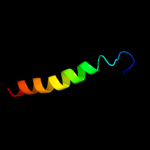
PDB 3npy chain B
Region: 173 - 185
Aligned: 13
Modelled: 13
Confidence: 11.0%
Identity: 38%
PDB header:oxidoreductase
Chain: B: PDB Molecule:tyrosinase;
PDBTitle: crystal structure of tyrosinase from bacillus megaterium soaked in2 cuso4
Phyre2
| 5 |
|
PDB 3qnq chain D
Region: 191 - 205
Aligned: 15
Modelled: 15
Confidence: 10.6%
Identity: 47%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 6 |
|
PDB 2y9x chain C
Region: 173 - 185
Aligned: 13
Modelled: 13
Confidence: 10.1%
Identity: 46%
PDB header:oxidoreductase
Chain: C: PDB Molecule:polyphenol oxidase;
PDBTitle: crystal structure of ppo3, a tyrosinase from agaricus bisporus, in2 deoxy-form that contains additional unknown lectin-like subunit,3 with inhibitor tropolone
Phyre2
| 7 |
|
PDB 3lk2 chain B
Region: 177 - 192
Aligned: 16
Modelled: 16
Confidence: 9.4%
Identity: 50%
PDB header:protein binding
Chain: B: PDB Molecule:f-actin-capping protein subunit beta isoforms 1 and 2;
PDBTitle: crystal structure of capz bound to the uncapping motif from carmil
Phyre2
| 8 |
|
PDB 1js8 chain A
Region: 173 - 185
Aligned: 13
Modelled: 13
Confidence: 9.3%
Identity: 38%
PDB header:oxygen storage/transport
Chain: A: PDB Molecule:hemocyanin;
PDBTitle: structure of a functional unit from octopus hemocyanin
Phyre2
| 9 |
|
PDB 1hf9 chain B
Region: 174 - 205
Aligned: 32
Modelled: 32
Confidence: 8.4%
Identity: 25%
PDB header:atpase inhibitor
Chain: B: PDB Molecule:atpase inhibitor (mitochondrial);
PDBTitle: c-terminal coiled-coil domain from bovine if1
Phyre2
| 10 |
|
PDB 1izn chain B
Region: 177 - 192
Aligned: 16
Modelled: 16
Confidence: 8.3%
Identity: 50%
Fold: Subunits of heterodimeric actin filament capping protein Capz
Superfamily: Subunits of heterodimeric actin filament capping protein Capz
Family: Capz beta-1 subunit
Phyre2
| 11 |
|
PDB 1js8 chain A domain 1
Region: 173 - 185
Aligned: 13
Modelled: 13
Confidence: 8.0%
Identity: 38%
Fold: Di-copper centre-containing domain
Superfamily: Di-copper centre-containing domain
Family: Hemocyanin middle domain
Phyre2
| 12 |
|
PDB 2k1k chain A
Region: 97 - 127
Aligned: 31
Modelled: 31
Confidence: 7.2%
Identity: 32%
PDB header:signaling protein
Chain: A: PDB Molecule:ephrin type-a receptor 1;
PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 4.3
Phyre2
| 13 |
|
PDB 2k1l chain B
Region: 97 - 127
Aligned: 31
Modelled: 31
Confidence: 7.2%
Identity: 32%
PDB header:signaling protein
Chain: B: PDB Molecule:ephrin type-a receptor 1;
PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 6.3
Phyre2
| 14 |
|
PDB 2k1k chain B
Region: 97 - 127
Aligned: 31
Modelled: 31
Confidence: 7.2%
Identity: 32%
PDB header:signaling protein
Chain: B: PDB Molecule:ephrin type-a receptor 1;
PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 4.3
Phyre2
| 15 |
|
PDB 2k1l chain A
Region: 97 - 127
Aligned: 31
Modelled: 31
Confidence: 7.2%
Identity: 32%
PDB header:signaling protein
Chain: A: PDB Molecule:ephrin type-a receptor 1;
PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 6.3
Phyre2
| 16 |
|
PDB 1wx2 chain A
Region: 173 - 185
Aligned: 13
Modelled: 13
Confidence: 7.2%
Identity: 31%
PDB header:oxidoreductase/metal transport
Chain: A: PDB Molecule:tyrosinase;
PDBTitle: crystal structure of the oxy-form of the copper-bound streptomyces2 castaneoglobisporus tyrosinase complexed with a caddie protein3 prepared by the addition of hydrogenperoxide
Phyre2
| 17 |
|
PDB 1u11 chain A
Region: 183 - 205
Aligned: 23
Modelled: 23
Confidence: 6.3%
Identity: 4%
Fold: Flavodoxin-like
Superfamily: N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family: N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Phyre2
| 18 |
|
PDB 2knc chain B
Region: 153 - 202
Aligned: 44
Modelled: 50
Confidence: 6.1%
Identity: 18%
PDB header:cell adhesion
Chain: B: PDB Molecule:integrin beta-3;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 19 |
|
PDB 2fqc chain A
Region: 92 - 98
Aligned: 7
Modelled: 7
Confidence: 5.8%
Identity: 43%
PDB header:toxin
Chain: A: PDB Molecule:conotoxin pl14a;
PDBTitle: solution structure of conotoxin pl14a
Phyre2
| 20 |
|
PDB 1lnl chain B
Region: 173 - 185
Aligned: 13
Modelled: 13
Confidence: 5.7%
Identity: 38%
PDB header:oxygen storage/transport
Chain: B: PDB Molecule:hemocyanin;
PDBTitle: structure of deoxygenated hemocyanin from rapana thomasiana
Phyre2
| 21 |
|
| 22 |
|