| 1 |
|
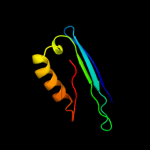
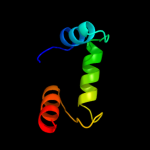
PDB 1mhm chain A
Region: 130 - 190
Aligned: 61
Modelled: 61
Confidence: 34.8%
Identity: 16%
PDB header:lyase
Chain: A: PDB Molecule:s-adenosylmethionine decarboxylase;
PDBTitle: crystal structure of s-adenosylmethionine decarboxylase2 from potato
Phyre2
| 2 |
|
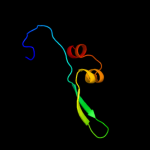
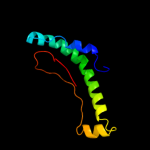
PDB 3ibp chain A
Region: 80 - 111
Aligned: 32
Modelled: 32
Confidence: 31.5%
Identity: 16%
PDB header:cell cycle
Chain: A: PDB Molecule:chromosome partition protein mukb;
PDBTitle: the crystal structure of the dimerization domain of escherichia coli2 structural maintenance of chromosomes protein mukb
Phyre2
| 3 |
|
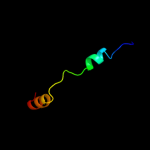
PDB 1msv chain B
Region: 130 - 190
Aligned: 58
Modelled: 61
Confidence: 28.3%
Identity: 12%
PDB header:lyase
Chain: B: PDB Molecule:s-adenosylmethionine decarboxylase proenzyme;
PDBTitle: the s68a s-adenosylmethionine decarboxylase proenzyme2 processing mutant.
Phyre2
| 4 |
|
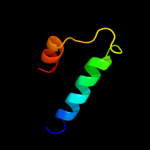
PDB 3ep3 chain A
Region: 130 - 190
Aligned: 58
Modelled: 61
Confidence: 23.2%
Identity: 12%
PDB header:lyase
Chain: A: PDB Molecule:s-adenosylmethionine decarboxylase alpha chain;
PDBTitle: human adometdc d174n mutant with no putrescine bound
Phyre2
| 5 |
|
PDB 1jl0 chain A
Region: 130 - 190
Aligned: 58
Modelled: 61
Confidence: 21.4%
Identity: 12%
Fold: S-adenosylmethionine decarboxylase
Superfamily: S-adenosylmethionine decarboxylase
Family: S-adenosylmethionine decarboxylase
Phyre2
| 6 |
|
PDB 1lva chain A domain 3
Region: 81 - 111
Aligned: 31
Modelled: 31
Confidence: 11.2%
Identity: 0%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: C-terminal fragment of elongation factor SelB
Phyre2
| 7 |
|
PDB 1l1o chain C
Region: 128 - 177
Aligned: 50
Modelled: 50
Confidence: 9.7%
Identity: 16%
Fold: OB-fold
Superfamily: Nucleic acid-binding proteins
Family: Single strand DNA-binding domain, SSB
Phyre2
| 8 |
|
PDB 3pcq chain M
Region: 1 - 28
Aligned: 28
Modelled: 28
Confidence: 8.1%
Identity: 32%
PDB header:photosynthesis
Chain: M: PDB Molecule:photosystem i reaction center subunit xii;
PDBTitle: femtosecond x-ray protein nanocrystallography
Phyre2
| 9 |
|
PDB 1mj5 chain A
Region: 171 - 185
Aligned: 15
Modelled: 15
Confidence: 7.5%
Identity: 20%
Fold: alpha/beta-Hydrolases
Superfamily: alpha/beta-Hydrolases
Family: Haloalkane dehalogenase
Phyre2
| 10 |
|
PDB 2wmm chain A
Region: 80 - 111
Aligned: 32
Modelled: 32
Confidence: 7.1%
Identity: 13%
PDB header:cell cycle
Chain: A: PDB Molecule:chromosome partition protein mukb;
PDBTitle: crystal structure of the hinge domain of mukb
Phyre2
| 11 |
|
PDB 1wsu chain A
Region: 76 - 110
Aligned: 34
Modelled: 35
Confidence: 5.8%
Identity: 12%
PDB header:translation/rna
Chain: A: PDB Molecule:selenocysteine-specific elongation factor;
PDBTitle: c-terminal domain of elongation factor selb complexed with2 secis rna
Phyre2
| 12 |
|
PDB 2q0o chain A
Region: 57 - 110
Aligned: 44
Modelled: 54
Confidence: 5.6%
Identity: 20%
PDB header:transcription
Chain: A: PDB Molecule:probable transcriptional activator protein trar;
PDBTitle: crystal structure of an anti-activation complex in bacterial quorum2 sensing
Phyre2
| 13 |
|
PDB 3mqz chain A
Region: 55 - 142
Aligned: 72
Modelled: 88
Confidence: 5.2%
Identity: 15%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized conserved protein duf1054;
PDBTitle: crystal structure of conserved protein duf1054 from pink subaerial2 biofilm microbial leptospirillum sp. group ii uba.
Phyre2