| 1 |
|
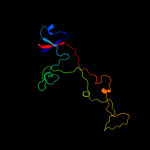
PDB 1pdi chain Q
Region: 7 - 165
Aligned: 159
Modelled: 159
Confidence: 100.0%
Identity: 25%
PDB header:structural protein
Chain: Q: PDB Molecule:short tail fiber protein;
PDBTitle: fitting of the c-terminal part of the short tail fibers2 into the cryo-em reconstruction of t4 baseplate
Phyre2
| 2 |
|
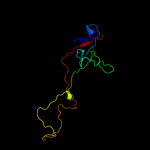
PDB 2xgf chain A
Region: 7 - 166
Aligned: 160
Modelled: 160
Confidence: 100.0%
Identity: 34%
PDB header:viral protein
Chain: A: PDB Molecule:long tail fiber protein p37;
PDBTitle: structure of the bacteriophage t4 long tail fibre needle-2 shaped receptor-binding tip
Phyre2
| 3 |
|
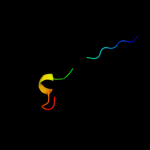
PDB 1ocy chain A
Region: 7 - 165
Aligned: 159
Modelled: 159
Confidence: 100.0%
Identity: 24%
Fold: Receptor-binding domain of short tail fibre protein gp12
Superfamily: Receptor-binding domain of short tail fibre protein gp12
Family: Receptor-binding domain of short tail fibre protein gp12
Phyre2
| 4 |
|
PDB 1h6w chain A
Region: 7 - 55
Aligned: 49
Modelled: 49
Confidence: 99.8%
Identity: 33%
PDB header:structural protein
Chain: A: PDB Molecule:bacteriophage t4 short tail fibre;
PDBTitle: crystal structure of a heat- and protease-stable fragment2 of the bacteriophage t4 short fibre
Phyre2
| 5 |
|
PDB 2fkk chain A
Region: 10 - 166
Aligned: 134
Modelled: 140
Confidence: 98.3%
Identity: 14%
PDB header:viral protein
Chain: A: PDB Molecule:baseplate structural protein gp10;
PDBTitle: crystal structure of the c-terminal domain of the bacteriophage t42 gene product 10
Phyre2
| 6 |
|
PDB 2fl8 chain N
Region: 10 - 166
Aligned: 134
Modelled: 140
Confidence: 98.2%
Identity: 14%
PDB header:virus/viral protein
Chain: N: PDB Molecule:baseplate structural protein gp10;
PDBTitle: fitting of the gp10 trimer structure into the cryoem map of the2 bacteriophage t4 baseplate in the hexagonal conformation.
Phyre2
| 7 |
|
PDB 1atx chain A
Region: 12 - 34
Aligned: 20
Modelled: 23
Confidence: 33.0%
Identity: 35%
Fold: Defensin-like
Superfamily: Defensin-like
Family: Defensin
Phyre2
| 8 |
|
PDB 1m5h chain A domain 2
Region: 12 - 57
Aligned: 40
Modelled: 46
Confidence: 7.3%
Identity: 25%
Fold: Ferredoxin-like
Superfamily: Formylmethanofuran:tetrahydromethanopterin formyltransferase
Family: Formylmethanofuran:tetrahydromethanopterin formyltransferase
Phyre2
| 9 |
|
PDB 1el6 chain A
Region: 3 - 25
Aligned: 23
Modelled: 23
Confidence: 6.2%
Identity: 22%
Fold: Baseplate structural protein gp11
Superfamily: Baseplate structural protein gp11
Family: Baseplate structural protein gp11
Phyre2
| 10 |
|
PDB 1s48 chain A
Region: 23 - 42
Aligned: 20
Modelled: 20
Confidence: 5.7%
Identity: 5%
Fold: DNA/RNA polymerases
Superfamily: DNA/RNA polymerases
Family: RNA-dependent RNA-polymerase
Phyre2
| 11 |
|
PDB 1v0d chain A
Region: 23 - 31
Aligned: 9
Modelled: 9
Confidence: 5.5%
Identity: 56%
PDB header:hydrolase
Chain: A: PDB Molecule:dna fragmentation factor 40 kda subunit;
PDBTitle: crystal structure of caspase-activated dnase (cad)
Phyre2
| 12 |
|
PDB 1v0d chain A
Region: 23 - 31
Aligned: 9
Modelled: 9
Confidence: 5.5%
Identity: 56%
Fold: His-Me finger endonucleases
Superfamily: His-Me finger endonucleases
Family: Caspase-activated DNase, CAD (DffB, DFF40)
Phyre2