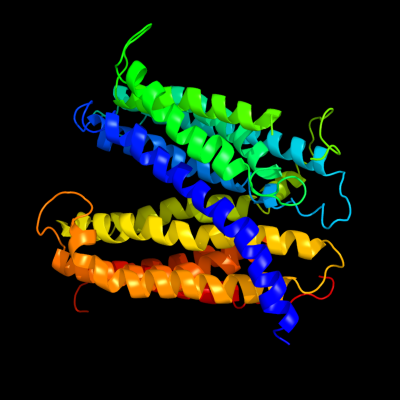
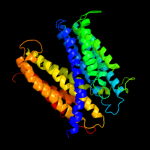
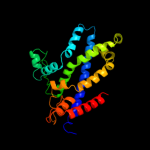
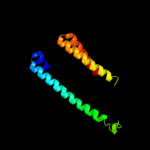
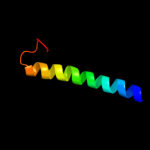
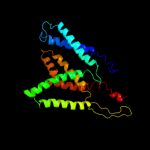
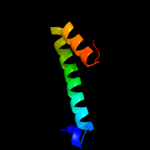
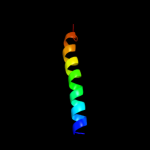1 c3mkuA_
100.0
19
PDB header: transport proteinChain: A: PDB Molecule: multi antimicrobial extrusion protein (na(+)/drugPDBTitle: structure of a cation-bound multidrug and toxin compound extrusion2 (mate) transporter
2 d2r6gf2
14.4
11
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like3 d1v54c_
13.9
9
Fold: Cytochrome c oxidase subunit III-likeSuperfamily: Cytochrome c oxidase subunit III-likeFamily: Cytochrome c oxidase subunit III-like4 d1u94a2
11.9
29
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain5 c1wrgA_
11.7
14
PDB header: membrane proteinChain: A: PDB Molecule: light-harvesting protein b-880, beta chain;PDBTitle: light-harvesting complex 1 beta subunit from wild-type2 rhodospirillum rubrum
6 c2yvxD_
10.8
13
PDB header: transport proteinChain: D: PDB Molecule: mg2+ transporter mgte;PDBTitle: crystal structure of magnesium transporter mgte
7 d1xp8a2
10.5
14
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain8 d1ubea2
10.4
43
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain9 d2yvxa3
10.4
13
Fold: MgtE membrane domain-likeSuperfamily: MgtE membrane domain-likeFamily: MgtE membrane domain-like10 d1lghb_
10.3
8
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits11 d1mo6a2
10.3
43
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain12 d2a5yb1
10.1
100
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CED-4 C-terminal domain-like13 d1sg7a1
10.0
8
Fold: ChaB-likeSuperfamily: ChaB-likeFamily: ChaB-like14 c1sg7A_
10.0
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative cation transport regulator chab;PDBTitle: nmr solution structure of the putative cation transport2 regulator chab
15 c3ipdB_
9.5
10
PDB header: exocytosisChain: B: PDB Molecule: syntaxin-1a;PDBTitle: helical extension of the neuronal snare complex into the2 membrane, spacegroup i 21 21 21
16 d1jo5a_
9.0
16
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits17 c2zqpY_
8.8
10
PDB header: protein transportChain: Y: PDB Molecule: preprotein translocase secy subunit;PDBTitle: crystal structure of secye translocon from thermus2 thermophilus
18 d2d5ba1
8.7
15
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases19 d2r6gg1
7.7
24
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like20 c1m57H_
7.4
24
PDB header: oxidoreductaseChain: H: PDB Molecule: cytochrome c oxidase;PDBTitle: structure of cytochrome c oxidase from rhodobacter2 sphaeroides (eq(i-286) mutant))
21 c1b9uA_
not modelled
7.3
10
PDB header: hydrolaseChain: A: PDB Molecule: protein (atp synthase);PDBTitle: membrane domain of the subunit b of the e.coli atp synthase
22 c2k1aA_
not modelled
7.2
14
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
23 c2l35A_
not modelled
7.1
18
PDB header: protein bindingChain: A: PDB Molecule: dap12-nkg2c_tm;PDBTitle: structure of the dap12-nkg2c transmembrane heterotrimer
24 c1cdlG_
not modelled
7.0
17
PDB header: calcium-binding proteinChain: G: PDB Molecule: calcium/calmodulin-dependent protein kinase typePDBTitle: target enzyme recognition by calmodulin: 2.4 angstroms2 structure of a calmodulin-peptide complex
25 c2o5gB_
not modelled
7.0
17
PDB header: metal binding proteinChain: B: PDB Molecule: smooth muscle myosin light chain kinase peptide;PDBTitle: calmodulin-smooth muscle light chain kinase peptide complex
26 c2ntxB_
not modelled
6.9
20
PDB header: signaling proteinChain: B: PDB Molecule: emb27 c1ygyA_
not modelled
6.9
18
PDB header: oxidoreductaseChain: A: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: crystal structure of d-3-phosphoglycerate dehydrogenase from2 mycobacterium tuberculosis
28 c1z65A_
not modelled
6.9
63
PDB header: unknown functionChain: A: PDB Molecule: prion-like protein doppel;PDBTitle: mouse doppel 1-30 peptide
29 d2hqxa1
not modelled
6.8
30
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: Tudor domain30 c2hqxB_
not modelled
6.8
30
PDB header: transcriptionChain: B: PDB Molecule: p100 co-activator tudor domain;PDBTitle: crystal structure of human p100 tudor domain conserved2 region
31 d3dtub2
not modelled
6.8
25
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region32 c3pnwX_
not modelled
6.8
13
PDB header: protein binding/immune systemChain: X: PDB Molecule: tudor domain-containing protein 3;PDBTitle: crystal structure of the tudor domain of human tdrd3 in complex with2 an anti-tdrd3 fab
33 c2eeyA_
not modelled
6.7
22
PDB header: biosynthetic proteinChain: A: PDB Molecule: molybdopterin biosynthesis;PDBTitle: structure of gk0241 protein from geobacillus kaustophilus
34 c2eqkA_
not modelled
6.7
14
PDB header: transcriptionChain: A: PDB Molecule: tudor domain-containing protein 4;PDBTitle: solution structure of the tudor domain of tudor domain-2 containing protein 4
35 d1nkzb_
not modelled
6.5
12
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits36 c2k9pA_
not modelled
6.4
18
PDB header: membrane proteinChain: A: PDB Molecule: pheromone alpha factor receptor;PDBTitle: structure of tm1_tm2 in lppg micelles
37 c2be6F_
not modelled
6.4
11
PDB header: membrane proteinChain: F: PDB Molecule: voltage-dependent l-type calcium channel alpha-1c subunit;PDBTitle: 2.0 a crystal structure of the cav1.2 iq domain-ca/cam complex
38 c1ql1A_
not modelled
6.4
24
PDB header: virusChain: A: PDB Molecule: pf1 bacteriophage coat protein b;PDBTitle: inovirus (filamentous bacteriophage) strain pf1 major coat2 protein assembly
39 d1u78a2
not modelled
6.2
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain40 d1mhna_
not modelled
6.2
18
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: Tudor domain41 d2hw4a1
not modelled
6.2
17
Fold: PHP14-likeSuperfamily: PHP14-likeFamily: Janus/Ocnus42 c3gg9C_
not modelled
6.2
17
PDB header: oxidoreductaseChain: C: PDB Molecule: d-3-phosphoglycerate dehydrogenase oxidoreductase protein;PDBTitle: crystal structure of putative d-3-phosphoglycerate dehydrogenase2 oxidoreductase from ralstonia solanacearum
43 c2hw4A_
not modelled
6.2
17
PDB header: structural genomics, hydrolaseChain: A: PDB Molecule: 14 kda phosphohistidine phosphatase;PDBTitle: crystal structure of human phosphohistidine phosphatase
44 c2d9tA_
not modelled
6.2
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: tudor domain-containing protein 3;PDBTitle: solution structure of the tudor domain of tudor domain2 containing protein 3 from mouse
45 c3egjA_
not modelled
6.1
25
PDB header: hydrolaseChain: A: PDB Molecule: n-acetylglucosamine-6-phosphate deacetylase;PDBTitle: n-acetylglucosamine-6-phosphate deacetylase from vibrio cholerae.
46 d1rqga1
not modelled
6.0
13
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases47 d1uptb_
not modelled
6.0
19
Fold: GRIP domainSuperfamily: GRIP domainFamily: GRIP domain48 d1u61a_
not modelled
5.9
19
Fold: RNase III domain-likeSuperfamily: RNase III domain-likeFamily: RNase III catalytic domain-like49 c1i97U_
not modelled
5.9
33
PDB header: ribosomeChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: crystal structure of the 30s ribosomal subunit from thermus2 thermophilus in complex with tetracycline
50 c2kvvA_
not modelled
5.9
33
PDB header: hydrolaseChain: A: PDB Molecule: putative excisionase;PDBTitle: solution nmr of putative excisionase from klebsiella pneumoniae,2 northeast structural genomics consortium target target kpr49
51 c3ci9B_
not modelled
5.8
15
PDB header: transcriptionChain: B: PDB Molecule: heat shock factor-binding protein 1;PDBTitle: crystal structure of the human hsbp1
52 c2b2aA_
not modelled
5.8
8
PDB header: transferaseChain: A: PDB Molecule: telomerase reverse transcriptase;PDBTitle: crystal structure of the ten domain of the telomerase2 reverse transcriptase
53 c1qfqB_
not modelled
5.8
42
PDB header: transcription/rnaChain: B: PDB Molecule: 36-mer n-terminal peptide of the n protein;PDBTitle: bacteriophage lambda n-protein-nutboxb-rna complex
54 c2fynH_
not modelled
5.8
19
PDB header: oxidoreductaseChain: H: PDB Molecule: cytochrome c1;PDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
55 c4a4fA_
not modelled
5.7
33
PDB header: rna binding proteinChain: A: PDB Molecule: survival of motor neuron-related-splicing factor 30;PDBTitle: solution structure of spf30 tudor domain in complex with2 symmetrically dimethylated arginine
56 c1g5vA_
not modelled
5.7
18
PDB header: translationChain: A: PDB Molecule: survival motor neuron protein 1;PDBTitle: solution structure of the tudor domain of the human smn2 protein
57 d1eysh2
not modelled
5.7
15
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction centre subunit H, transmembrane regionFamily: Photosystem II reaction centre subunit H, transmembrane region58 d1m56d_
not modelled
5.6
11
Fold: Single transmembrane helixSuperfamily: Bacterial aa3 type cytochrome c oxidase subunit IVFamily: Bacterial aa3 type cytochrome c oxidase subunit IV59 c3mp7B_
not modelled
5.6
18
PDB header: protein transportChain: B: PDB Molecule: preprotein translocase subunit sece;PDBTitle: lateral opening of a translocon upon entry of protein suggests the2 mechanism of insertion into membranes
60 d1ik0a_
not modelled
5.6
21
Fold: 4-helical cytokinesSuperfamily: 4-helical cytokinesFamily: Short-chain cytokines61 c3s6n2_
not modelled
5.6
8
PDB header: splicingChain: 2: PDB Molecule: survival of motor neuron protein-interacting protein 1;PDB Fragment: gemin2-binding domain;
PDBTitle: crystal structure of the gemin2-binding domain of smn, gemin2 in2 complex with smd1/d2/f/e/g from human
62 c3txsC_
not modelled
5.6
21
PDB header: viral proteinChain: C: PDB Molecule: terminase dna packaging enzyme small subunit;PDBTitle: crystal structure of phage 44rr small terminase gp16
63 d3b60a2
not modelled
5.6
9
Fold: ABC transporter transmembrane regionSuperfamily: ABC transporter transmembrane regionFamily: ABC transporter transmembrane region64 c2ow8v_
not modelled
5.5
33
PDB header: ribosomeChain: V: PDB Molecule: PDBTitle: crystal structure of a 70s ribosome-trna complex reveals functional2 interactions and rearrangements. this file, 2ow8, contains the 30s3 ribosome subunit, two trna, and mrna molecules. 50s ribosome subunit4 is in the file 1vsa.
65 c2dbqA_
not modelled
5.5
24
PDB header: oxidoreductaseChain: A: PDB Molecule: glyoxylate reductase;PDBTitle: crystal structure of glyoxylate reductase (ph0597) from pyrococcus2 horikoshii ot3, complexed with nadp (i41)
66 d2d9ta1
not modelled
5.5
13
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: Tudor domain67 c3kf9B_
not modelled
5.4
33
PDB header: cell cycle/calcium-binding proteinChain: B: PDB Molecule: myosin light chain kinase 2, skeletal/cardiac muscle;PDBTitle: crystal structure of the sdcen/skmlck complex
68 c2hydB_
not modelled
5.4
8
PDB header: transport proteinChain: B: PDB Molecule: abc transporter homolog;PDBTitle: multidrug abc transporter sav1866
69 c3bbo2_
not modelled
5.4
15
PDB header: ribosomeChain: 2: PDB Molecule: ribosomal protein l32;PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
70 d1d8ja_
not modelled
5.4
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: The central core domain of TFIIE beta71 c2yggA_
not modelled
5.4
17
PDB header: metal binding protein/transport proteinChain: A: PDB Molecule: sodium/hydrogen exchanger 1;PDBTitle: complex of cambr and cam
72 c2uxbU_
not modelled
5.3
33
PDB header: ribosomeChain: U: PDB Molecule: ribosomal protein thx;PDBTitle: crystal structure of an extended trna anticodon stem loop2 in complex with its cognate mrna gggu in the context of3 the thermus thermophilus 30s subunit.
73 c2uuaU_
not modelled
5.3
33
PDB header: ribosomeChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: structure of the thermus thermophilus 30s ribosomal subunit2 complexed with a valine-asl with cmo5u in position 343 bound to an mrna with a guc-codon in the a-site and4 paromomycin.
74 d2gu3a1
not modelled
5.3
11
Fold: Cystatin-likeSuperfamily: Cystatin/monellinFamily: PepSY-like75 c2j02U_
not modelled
5.3
33
PDB header: ribosomeChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: structure of the thermus thermophilus 70s ribosome2 complexed with mrna, trna and paromomycin (part 3 of 4)3 this file contains the 30s subunit, mrna, a-, p- and4 e-site trnas and paromomycin for molecule ii.
76 c1hnwV_
not modelled
5.3
33
PDB header: ribosomeChain: V: PDB Molecule: 30s ribosomal protein thx;PDBTitle: structure of the thermus thermophilus 30s ribosomal subunit2 in complex with tetracycline
77 c1j5eV_
not modelled
5.3
33
PDB header: ribosomeChain: V: PDB Molecule: 30s ribosomal protein thx;PDBTitle: structure of the thermus thermophilus 30s ribosomal subunit
78 c2uxcU_
not modelled
5.3
33
PDB header: ribosomeChain: U: PDB Molecule: ribosomal protein thx;PDBTitle: crystal structure of an extended trna anticodon stem loop2 in complex with its cognate mrna ucgu in the context of3 the thermus thermophilus 30s subunit.
79 c3ficU_
not modelled
5.3
33
PDB header: ribosomeChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: t. thermophilus 70s ribosome in complex with mrna, trnas and ef-2 tu.gdp.kirromycin ternary complex, fitted to a 6.4 a cryo-em map.3 this file contains the 30s subunit and the ligands
80 c1xmqV_
not modelled
5.3
33
PDB header: ribosomeChain: V: PDB Molecule: 30s ribosomal protein thx;PDBTitle: crystal structure of t6a37-asllysuuu aaa-mrna bound to the decoding2 center
81 c2uucU_
not modelled
5.3
33
PDB header: ribosomeChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: structure of the thermus thermophilus 30s ribosomal subunit2 complexed with a valine-asl with cmo5u in position 343 bound to an mrna with a gua-codon in the a-site and4 paromomycin.
82 c2vqfU_
not modelled
5.3
33
PDB header: ribosomeChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: modified uridines with c5-methylene substituents at the2 first position of the trna anticodon stabilize u-g wobble3 pairing during decoding
83 c1ibkV_
not modelled
5.3
33
PDB header: ribosomeChain: V: PDB Molecule: 30s ribosomal protein thx;PDBTitle: structure of the thermus thermophilus 30s ribosomal subunit2 in complex with the antibiotic paromomycin
84 c1hr0V_
not modelled
5.3
33
PDB header: ribosomeChain: V: PDB Molecule: 30s ribosomal protein thx;PDBTitle: crystal structure of initiation factor if1 bound to the 30s2 ribosomal subunit
85 c1hnzV_
not modelled
5.3
33
PDB header: ribosomeChain: V: PDB Molecule: 30s ribosomal protein thx;PDBTitle: structure of the thermus thermophilus 30s ribosomal subunit2 in complex with hygromycin b
86 c1n32V_
not modelled
5.3
33
PDB header: ribosomeChain: V: PDB Molecule: 30s ribosomal protein thx;PDBTitle: structure of the thermus thermophilus 30s ribosomal subunit2 bound to codon and near-cognate transfer rna anticodon3 stem-loop mismatched at the first codon position at the a4 site with paromomycin
87 c2uubU_
not modelled
5.3
33
PDB header: ribosomeChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: structure of the thermus thermophilus 30s ribosomal subunit2 complexed with a valine-asl with cmo5u in position 343 bound to an mrna with a guu-codon in the a-site and4 paromomycin.
88 c3zvoU_
not modelled
5.3
33
PDB header: ribosomeChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: crystal structure of the hybrid state of ribosome in complex with2 the guanosine triphosphatase release factor 3
89 c3oi4U_
not modelled
5.3
33
PDB header: ribosome/antibioticChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: structure of the thermus thermophilus 70s ribosome complexed with2 telithromycin. this file contains the 30s subunit of one 70s3 ribosome. the entire crystal structure contains two 70s ribosomes.
90 c3oi2U_
not modelled
5.3
33
PDB header: ribosome/antibioticChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: structure of the thermus thermophilus 70s ribosome complexed with2 telithromycin. this file contains the 30s subunit of one 70s3 ribosome. the entire crystal structure contains two 70s ribosomes.
91 c3mr8U_
not modelled
5.3
33
PDB header: ribosomeChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: recognition of the amber stop codon by release factor rf1. this entry2 3mr8 contains 30s ribosomal subunit. the 50s ribosomal subunit can be3 found in pdb entry 3ms1. molecule b in the same asymmetric unit is4 deposited as 3mrz (50s) and 3ms0 (30s).
92 c2j00U_
not modelled
5.3
33
PDB header: ribosomeChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: structure of the thermus thermophilus 70s ribosome2 complexed with mrna, trna and paromomycin (part 1 of 4).3 this file contains the 30s subunit, mrna, a-, p- and4 e-site trnas and paromomycin for molecule i.
93 c2y12U_
not modelled
5.3
33
PDB header: ribosomeChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: the crystal structure of ef-tu and g24a-trna-trp bound to a2 near-cognate codon on the 70s ribosome
94 c2vqeU_
not modelled
5.3
33
PDB header: ribosomeChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: modified uridines with c5-methylene substituents at the2 first position of the trna anticodon stabilize u-g wobble3 pairing during decoding
95 c3ms0U_
not modelled
5.3
33
PDB header: ribosomeChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: recognition of the amber stop codon by release factor rf1. this entry2 3ms0 contains 30s ribosomal subunit. the 50s ribosomal subunit can be3 found in pdb entry 3mrz. molecule a in the same asymmetric unit is4 deposited as 3mr8 (30s) and 3ms1 (50s).
96 c1ar1B_
not modelled
5.3
25
PDB header: complex (oxidoreductase/antibody)Chain: B: PDB Molecule: cytochrome c oxidase;PDBTitle: structure at 2.7 angstrom resolution of the paracoccus2 denitrificans two-subunit cytochrome c oxidase complexed3 with an antibody fv fragment
97 c1qleB_
not modelled
5.3
25
PDB header: oxidoreductase/immune systemChain: B: PDB Molecule: cytochrome c oxidase polypeptide ii;PDBTitle: cryo-structure of the paracoccus denitrificans four-subunit2 cytochrome c oxidase in the completely oxidized state3 complexed with an antibody fv fragment
98 c3ogeU_
not modelled
5.3
33
PDB header: ribosome/antibioticChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: structure of the thermus thermophilus ribosome complexed with2 chloramphenicol. this file contains the 30s subunit of one 70s3 ribosome. the entire crystal structure contains two 70s ribosomes.
99 c1gixX_
not modelled
5.3
33
PDB header: ribosomeChain: X: PDB Molecule: 30s ribosomal protein thx;PDBTitle: crystal structure of the ribosome at 5.5 a resolution. this file,2 1gix, contains the 30s ribosome subunit, three trna, and mrna3 molecules. 50s ribosome subunit is in the file 1giy