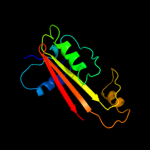
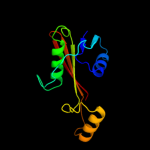
1 d1ex2a_
100.0
48
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: Maf-like2 d2amha1
100.0
20
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: Maf-like3 c2p5xB_
100.0
38
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: n-acetylserotonin o-methyltransferase-like protein;PDBTitle: crystal structure of maf domain of human n-acetylserotonin o-2 methyltransferase-like protein
4 d1v7ra_
96.7
11
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: ITPase (Ham1)5 d2cara1
95.5
23
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: ITPase (Ham1)6 d1k7ka_
95.1
17
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: ITPase (Ham1)7 d1b78a_
92.5
19
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: ITPase (Ham1)8 c3tquD_
90.0
22
PDB header: hydrolaseChain: D: PDB Molecule: non-canonical purine ntp pyrophosphatase;PDBTitle: structure of a ham1 protein from coxiella burnetii
9 d1vp2a_
89.6
15
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: ITPase (Ham1)10 d1pdaa1
63.7
25
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like11 c2ypnA_
62.4
25
PDB header: transferaseChain: A: PDB Molecule: protein (hydroxymethylbilane synthase);PDBTitle: hydroxymethylbilane synthase
12 c3eq1A_
61.0
21
PDB header: transferaseChain: A: PDB Molecule: porphobilinogen deaminase;PDBTitle: the crystal structure of human porphobilinogen deaminase at2 2.8a resolution
13 c2db5A_
40.3
19
PDB header: protein bindingChain: A: PDB Molecule: inad-like protein;PDBTitle: solution structure of the first pdz domain of inad-like2 protein
14 c2o2tB_
38.4
24
PDB header: structural proteinChain: B: PDB Molecule: multiple pdz domain protein;PDBTitle: the crystal structure of the 1st pdz domain of mpdz
15 c2e7kA_
32.4
26
PDB header: membrane proteinChain: A: PDB Molecule: maguk p55 subfamily member 2;PDBTitle: solution structure of the pdz domain from human maguk p552 subfamily member 2
16 c1x3lA_
31.2
22
PDB header: transferaseChain: A: PDB Molecule: hypothetical protein ph0495;PDBTitle: crystal structure of the ph0495 protein from pyrococccus horikoshii2 ot3
17 c3eggC_
29.8
25
PDB header: hydrolaseChain: C: PDB Molecule: spinophilin;PDBTitle: crystal structure of a complex between protein phosphatase 1 alpha2 (pp1) and the pp1 binding and pdz domains of spinophilin
18 c3jr2D_
29.2
11
PDB header: biosynthetic proteinChain: D: PDB Molecule: hexulose-6-phosphate synthase sgbh;PDBTitle: x-ray crystal structure of the mg-bound 3-keto-l-gulonate-6-phosphate2 decarboxylase from vibrio cholerae o1 biovar el tor str. n16961
19 c3a11D_
28.8
25
PDB header: isomeraseChain: D: PDB Molecule: translation initiation factor eif-2b, deltaPDBTitle: crystal structure of ribose-1,5-bisphosphate isomerase from2 thermococcus kodakaraensis kod1
20 d1t5oa_
23.0
19
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: IF2B-like21 d1ydua1
not modelled
21.8
20
Fold: At5g01610-likeSuperfamily: At5g01610-likeFamily: At5g01610-like22 d1gh9a_
not modelled
19.3
23
Fold: Rubredoxin-likeSuperfamily: Hypothetical protein MTH1184Family: Hypothetical protein MTH118423 d1wh1a_
not modelled
18.8
21
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain24 d1y0ua_
not modelled
18.4
33
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ArsR-like transcriptional regulators25 d1v6ba_
not modelled
17.9
23
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain26 d1a9xa4
not modelled
17.4
13
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like27 d1ujva_
not modelled
17.2
17
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain28 c1tuoA_
not modelled
17.2
11
PDB header: biosynthetic proteinChain: A: PDB Molecule: putative phosphomannomutase;PDBTitle: crystal structure of putative phosphomannomutase from2 thermus thermophilus hb8
29 d2eyqa2
not modelled
15.8
29
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain30 c2x0kB_
not modelled
14.7
18
PDB header: transferaseChain: B: PDB Molecule: riboflavin biosynthesis protein ribf;PDBTitle: crystal structure of modular fad synthetase from2 corynebacterium ammoniagenes
31 d1i16a_
not modelled
14.5
17
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: Interleukin 1632 c3hrdC_
not modelled
13.1
15
PDB header: oxidoreductaseChain: C: PDB Molecule: nicotinate dehydrogenase fad-subunit;PDBTitle: crystal structure of nicotinate dehydrogenase
33 d1gpua2
not modelled
12.1
16
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: TK-like Pyr module34 d1vaea_
not modelled
12.1
18
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain35 d1e94a_
not modelled
12.0
15
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits36 d1sknp_
not modelled
11.9
23
Fold: A DNA-binding domain in eukaryotic transcription factorsSuperfamily: A DNA-binding domain in eukaryotic transcription factorsFamily: A DNA-binding domain in eukaryotic transcription factors37 d1jgta2
not modelled
11.7
32
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Class II glutamine amidotransferases38 d1ujda_
not modelled
11.7
17
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain39 c2o7pA_
not modelled
11.6
14
PDB header: hydrolase, oxidoreductaseChain: A: PDB Molecule: riboflavin biosynthesis protein ribd;PDBTitle: the crystal structure of ribd from escherichia coli in complex with2 the oxidised nadp+ cofactor in the active site of the reductase3 domain
40 d1nd9a_
not modelled
11.4
7
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: N-terminal subdomain of bacterial translation initiation factor IF241 d1t1va_
not modelled
10.6
18
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: SH3BGR (SH3-binding, glutamic acid-rich protein-like)42 d1vb5a_
not modelled
10.4
20
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: IF2B-like43 c3diwB_
not modelled
10.2
23
PDB header: signaling protein/cell adhesionChain: B: PDB Molecule: tax1-binding protein 3;PDBTitle: c-terminal beta-catenin bound tip-1 structure
44 c1u37A_
not modelled
9.8
19
PDB header: protein transportChain: A: PDB Molecule: amyloid beta a4 precursor protein-binding,PDBTitle: auto-inhibition mechanism of x11s/mints family scaffold2 proteins revealed by the closed conformation of the tandem3 pdz domains
45 c1u38A_
not modelled
9.8
19
PDB header: protein transportChain: A: PDB Molecule: amyloid beta a4 precursor protein-binding,PDBTitle: auto-inhibition mechanism of x11s/mints family scaffold2 proteins revealed by the closed conformation of the tandem3 pdz domains
46 d1rp5a2
not modelled
9.8
11
Fold: Penicillin-binding protein 2x (pbp-2x), c-terminal domainSuperfamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domainFamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domain47 c2edpA_
not modelled
9.7
18
PDB header: structural proteinChain: A: PDB Molecule: shroom family member 4;PDBTitle: solution structure of the pdz domain from human shroom2 family member 4
48 d1kwma2
not modelled
9.7
10
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Pancreatic carboxypeptidase, activation domain49 d1x4pa1
not modelled
9.6
28
Fold: Surp module (SWAP domain)Superfamily: Surp module (SWAP domain)Family: Surp module (SWAP domain)50 d1u5wa1
not modelled
9.6
10
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: YjjX-like51 d1id3b_
not modelled
9.6
12
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones52 c2ehrA_
not modelled
9.5
14
PDB header: structural proteinChain: A: PDB Molecule: inad-like protein;PDBTitle: solution structure of the sixth pdz domain of human inad-2 like protein
53 c1m1zB_
not modelled
9.5
43
PDB header: hydrolaseChain: B: PDB Molecule: beta-lactam synthetase;PDBTitle: beta-lactam synthetase apo enzyme
54 d1f2ea2
not modelled
9.3
18
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione S-transferase (GST), N-terminal domain55 c3b76A_
not modelled
9.3
17
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase lnx;PDBTitle: crystal structure of the third pdz domain of human ligand-of-numb2 protein-x (lnx1) in complex with the c-terminal peptide from the3 coxsackievirus and adenovirus receptor
56 c3cyyA_
not modelled
9.3
22
PDB header: peptide binding proteinChain: A: PDB Molecule: tight junction protein zo-1;PDBTitle: the crystal structure of zo-1 pdz2 in complex with the cx43 peptide
57 c2ktaA_
not modelled
9.1
17
PDB header: hydrolaseChain: A: PDB Molecule: putative helicase;PDBTitle: solution nmr structure of a domain of protein a6ky75 from bacteroides2 vulgatus, northeast structural genomics target bvr106a
58 d1ayea2
not modelled
9.1
10
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Pancreatic carboxypeptidase, activation domain59 d2boaa2
not modelled
9.1
10
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Pancreatic carboxypeptidase, activation domain60 d1p5dx2
not modelled
9.1
15
Fold: Phosphoglucomutase, first 3 domainsSuperfamily: Phosphoglucomutase, first 3 domainsFamily: Phosphoglucomutase, first 3 domains61 c1q5rD_
not modelled
8.8
20
PDB header: hydrolaseChain: D: PDB Molecule: proteasome alpha-type subunit 1;PDBTitle: the rhodococcus 20s proteasome with unprocessed pro-peptides
62 d1v8aa_
not modelled
8.8
13
Fold: Ribokinase-likeSuperfamily: Ribokinase-likeFamily: Thiamin biosynthesis kinases63 d2cupa1
not modelled
8.7
29
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain64 c2kz5A_
not modelled
8.7
27
PDB header: transcriptionChain: A: PDB Molecule: transcription factor nf-e2 45 kda subunit;PDBTitle: solution nmr structure of transcription factor nf-e2 subunit's dna2 binding domain from homo sapiens, northeast structural genomics3 consortium target hr4653b
65 c2yt7A_
not modelled
8.7
19
PDB header: protein transportChain: A: PDB Molecule: amyloid beta a4 precursor protein-binding familyPDBTitle: solution structure of the pdz domain of amyloid beta a42 precursor protein-binding family a member 3
66 d2h3la1
not modelled
8.7
21
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain67 c2dluA_
not modelled
8.7
23
PDB header: protein bindingChain: A: PDB Molecule: inad-like protein;PDBTitle: solution structure of the second pdz domain of human inad-2 like protein
68 c2cveA_
not modelled
8.6
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ttha1053;PDBTitle: crystal structure of a conserved hypothetical protein tt1547 from2 thermus thermophilus hb8
69 d2z3ba1
not modelled
8.5
23
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits70 d1x63a1
not modelled
8.5
29
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain71 c3qglD_
not modelled
8.4
18
PDB header: protein bindingChain: D: PDB Molecule: sorting nexin-27;PDBTitle: crystal structure of pdz domain of sorting nexin 27 (snx27) in complex2 with the eseskv peptide corresponding to the c-terminal tail of girk3
72 c2krgA_
not modelled
8.3
23
PDB header: signaling proteinChain: A: PDB Molecule: na(+)/h(+) exchange regulatory cofactor nhe-rf1;PDBTitle: solution structure of human sodium/ hydrogen exchange2 regulatory factor 1(150-358)
73 d1v5qa_
not modelled
8.3
29
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain74 c1v5qA_
not modelled
8.3
29
PDB header: protein bindingChain: A: PDB Molecule: glutamate receptor interacting protein 1a-lPDBTitle: solution structure of the pdz domain from mouse glutamate2 receptor interacting protein 1a-l (grip1) homolog
75 d1x5ra1
not modelled
8.2
28
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain76 d1pyta_
not modelled
8.2
10
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Pancreatic carboxypeptidase, activation domain77 d1k25a2
not modelled
8.1
5
Fold: Penicillin-binding protein 2x (pbp-2x), c-terminal domainSuperfamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domainFamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domain78 d1g3ka_
not modelled
8.1
23
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits79 d1b65a_
not modelled
8.1
14
Fold: DmpA/ArgJ-likeSuperfamily: DmpA/ArgJ-likeFamily: DmpA-like80 c3dzvB_
not modelled
8.1
8
PDB header: transferaseChain: B: PDB Molecule: 4-methyl-5-(beta-hydroxyethyl)thiazole kinase;PDBTitle: crystal structure of 4-methyl-5-(beta-hydroxyethyl)thiazole2 kinase (np_816404.1) from enterococcus faecalis v583 at3 2.57 a resolution
81 d1q7xa_
not modelled
8.1
29
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain82 d1pyya2
not modelled
8.0
16
Fold: Penicillin-binding protein 2x (pbp-2x), c-terminal domainSuperfamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domainFamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domain83 d1itza2
not modelled
8.0
16
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: TK-like Pyr module84 c2du4B_
not modelled
8.0
21
PDB header: ligase/rnaChain: B: PDB Molecule: o-phosphoseryl-trna synthetase;PDBTitle: crystal structure of archaeoglobus fulgidus o-phosphoseryl-2 trna synthetase complexed with trnacys
85 d1obxa_
not modelled
7.9
14
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain86 c1obxA_
not modelled
7.9
14
PDB header: cell adhesionChain: A: PDB Molecule: syntenin 1;PDBTitle: crystal structure of the complex of pdz2 of syntenin with2 an interleukin 5 receptor alpha peptide.
87 c1obyB_
not modelled
7.9
14
PDB header: cell adhesionChain: B: PDB Molecule: syntenin 1;PDBTitle: crystal structure of the complex of pdz2 of syntenin with2 a syndecan-4 peptide.
88 d1nsaa2
not modelled
7.9
15
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Pancreatic carboxypeptidase, activation domain89 c3lyvF_
not modelled
7.8
40
PDB header: chaperoneChain: F: PDB Molecule: ribosome-associated factor y;PDBTitle: crystal structure of a domain of ribosome-associated factor y from2 streptococcus pyogenes serotype m6. northeast structural genomics3 consortium target id dr64a
90 c2ejyA_
not modelled
7.7
14
PDB header: membrane proteinChain: A: PDB Molecule: 55 kda erythrocyte membrane protein;PDBTitle: solution structure of the p55 pdz t85c domain complexed2 with the glycophorin c f127c peptide
91 d1m5za_
not modelled
7.7
11
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain92 d1pcaa1
not modelled
7.7
10
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Pancreatic carboxypeptidase, activation domain93 c3gkxB_
not modelled
7.7
10
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative arsc family related protein;PDBTitle: crystal structure of putative arsc family related protein from2 bacteroides fragilis
94 c2iwqA_
not modelled
7.7
26
PDB header: signaling proteinChain: A: PDB Molecule: multiple pdz domain protein;PDBTitle: 7th pdz domain of multiple pdz domain protein mpdz
95 d1um7a_
not modelled
7.7
32
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain96 d1ueqa_
not modelled
7.6
17
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain97 d1mfga_
not modelled
7.6
21
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain98 d2a0ua1
not modelled
7.6
27
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: IF2B-like99 d2dlda2
not modelled
7.5
8
Fold: Flavodoxin-likeSuperfamily: Formate/glycerate dehydrogenase catalytic domain-likeFamily: Formate/glycerate dehydrogenases, substrate-binding domain