| 1 |
|
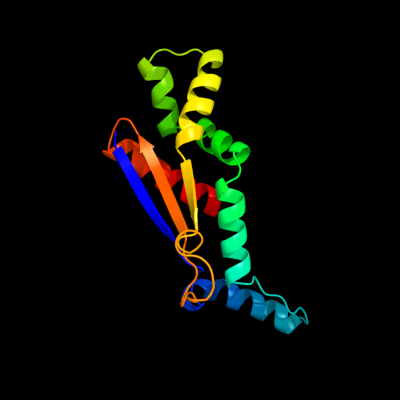
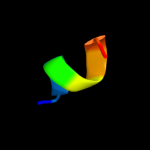
PDB 3b8m chain A domain 1
Region: 10 - 143
Aligned: 128
Modelled: 134
Confidence: 100.0%
Identity: 98%
Fold: Ferredoxin-like
Superfamily: Bacterial polysaccharide co-polymerase-like
Family: FepE-like
Phyre2
| 2 |
|
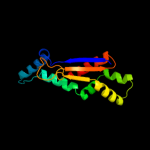
PDB 3b8o chain A domain 1
Region: 10 - 143
Aligned: 122
Modelled: 128
Confidence: 100.0%
Identity: 16%
Fold: Ferredoxin-like
Superfamily: Bacterial polysaccharide co-polymerase-like
Family: FepE-like
Phyre2
| 3 |
|
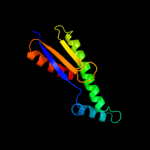
PDB 3b8p chain A domain 1
Region: 9 - 143
Aligned: 104
Modelled: 104
Confidence: 99.9%
Identity: 24%
Fold: Ferredoxin-like
Superfamily: Bacterial polysaccharide co-polymerase-like
Family: FepE-like
Phyre2
| 4 |
|
PDB 1p5d chain X domain 4
Region: 118 - 144
Aligned: 27
Modelled: 27
Confidence: 21.2%
Identity: 22%
Fold: TBP-like
Superfamily: Phosphoglucomutase, C-terminal domain
Family: Phosphoglucomutase, C-terminal domain
Phyre2
| 5 |
|
PDB 1mix chain A domain 2
Region: 119 - 141
Aligned: 23
Modelled: 23
Confidence: 17.2%
Identity: 30%
Fold: PH domain-like barrel
Superfamily: PH domain-like
Family: Third domain of FERM
Phyre2
| 6 |
|
PDB 1mji chain A
Region: 117 - 137
Aligned: 21
Modelled: 21
Confidence: 11.2%
Identity: 33%
Fold: RL5-like
Superfamily: RL5-like
Family: Ribosomal protein L5
Phyre2
| 7 |
|
PDB 3bbo chain H
Region: 117 - 137
Aligned: 21
Modelled: 21
Confidence: 10.0%
Identity: 33%
PDB header:ribosome
Chain: H: PDB Molecule:ribosomal protein l5;
PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
Phyre2
| 8 |
|
PDB 2zjr chain D domain 1
Region: 117 - 137
Aligned: 21
Modelled: 21
Confidence: 10.0%
Identity: 38%
Fold: RL5-like
Superfamily: RL5-like
Family: Ribosomal protein L5
Phyre2
| 9 |
|
PDB 1mk7 chain B
Region: 119 - 141
Aligned: 23
Modelled: 23
Confidence: 9.9%
Identity: 30%
PDB header:structural protein
Chain: B: PDB Molecule:talin;
PDBTitle: crystal structure of an integrin beta3-talin chimera
Phyre2
| 10 |
|
PDB 1iq4 chain A
Region: 117 - 137
Aligned: 21
Modelled: 21
Confidence: 8.2%
Identity: 33%
Fold: RL5-like
Superfamily: RL5-like
Family: Ribosomal protein L5
Phyre2
| 11 |
|
PDB 2k9i chain B
Region: 3 - 11
Aligned: 9
Modelled: 9
Confidence: 7.7%
Identity: 33%
PDB header:dna binding protein
Chain: B: PDB Molecule:plasmid prn1, complete sequence;
PDBTitle: nmr structure of plasmid copy control protein orf56 from2 sulfolobus islandicus
Phyre2
| 12 |
|
PDB 2jvf chain A
Region: 82 - 139
Aligned: 46
Modelled: 58
Confidence: 7.6%
Identity: 17%
PDB header:de novo protein
Chain: A: PDB Molecule:de novo protein m7;
PDBTitle: solution structure of m7, a computationally-designed2 artificial protein
Phyre2
| 13 |
|
PDB 3g9w chain A
Region: 119 - 141
Aligned: 23
Modelled: 23
Confidence: 7.2%
Identity: 30%
PDB header:cell adhesion
Chain: A: PDB Molecule:talin-2;
PDBTitle: crystal structure of talin2 f2-f3 in complex with the integrin beta1d2 cytoplasmic tail
Phyre2
| 14 |
|
PDB 1eik chain A
Region: 126 - 137
Aligned: 12
Modelled: 12
Confidence: 7.1%
Identity: 33%
Fold: RPB5-like RNA polymerase subunit
Superfamily: RPB5-like RNA polymerase subunit
Family: RPB5
Phyre2
| 15 |
|
PDB 1dzf chain A domain 2
Region: 126 - 137
Aligned: 12
Modelled: 12
Confidence: 6.9%
Identity: 33%
Fold: RPB5-like RNA polymerase subunit
Superfamily: RPB5-like RNA polymerase subunit
Family: RPB5
Phyre2
| 16 |
|
PDB 2pmz chain V
Region: 126 - 137
Aligned: 12
Modelled: 12
Confidence: 6.9%
Identity: 25%
PDB header:translation, transferase
Chain: V: PDB Molecule:dna-directed rna polymerase subunit h;
PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
Phyre2
| 17 |
|
PDB 1hmj chain A
Region: 126 - 137
Aligned: 12
Modelled: 12
Confidence: 6.3%
Identity: 33%
Fold: RPB5-like RNA polymerase subunit
Superfamily: RPB5-like RNA polymerase subunit
Family: RPB5
Phyre2
| 18 |
|
PDB 2a5y chain B domain 2
Region: 132 - 141
Aligned: 10
Modelled: 10
Confidence: 6.2%
Identity: 40%
Fold: DEATH domain
Superfamily: DEATH domain
Family: Caspase recruitment domain, CARD
Phyre2
| 19 |
|
PDB 3n7n chain E
Region: 41 - 55
Aligned: 15
Modelled: 15
Confidence: 6.1%
Identity: 20%
PDB header:replication
Chain: E: PDB Molecule:monopolin complex subunit lrs4;
PDBTitle: structure of csm1/lrs4 complex
Phyre2
| 20 |
|
PDB 1uuj chain A
Region: 6 - 13
Aligned: 8
Modelled: 8
Confidence: 6.0%
Identity: 63%
Fold: Lissencephaly-1 protein (Lis-1, PAF-AH alpha) N-terminal domain
Superfamily: Lissencephaly-1 protein (Lis-1, PAF-AH alpha) N-terminal domain
Family: Lissencephaly-1 protein (Lis-1, PAF-AH alpha) N-terminal domain
Phyre2
| 21 |
|