| 1 |
|
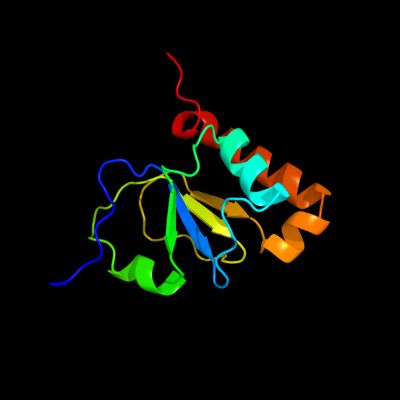
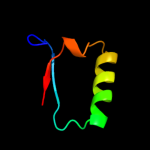
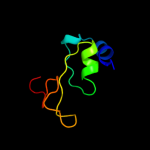
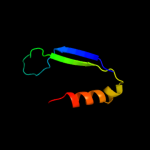
PDB 1em8 chain B
Region: 27 - 136
Aligned: 110
Modelled: 110
Confidence: 100.0%
Identity: 100%
Fold: DNA polymerase III psi subunit
Superfamily: DNA polymerase III psi subunit
Family: DNA polymerase III psi subunit
Phyre2
| 2 |
|
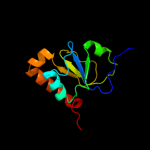
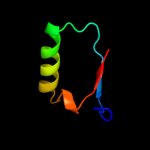
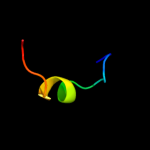
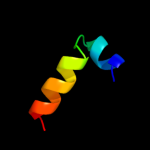
PDB 3sxu chain B
Region: 33 - 134
Aligned: 102
Modelled: 102
Confidence: 100.0%
Identity: 100%
PDB header:transferase
Chain: B: PDB Molecule:dna polymerase iii subunit psi;
PDBTitle: structure of the e. coli ssb-dna polymerase iii interface
Phyre2
| 3 |
|
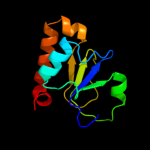
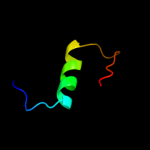
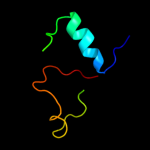
PDB 3gli chain P
Region: 2 - 28
Aligned: 27
Modelled: 27
Confidence: 98.4%
Identity: 100%
PDB header:transferase/dna
Chain: P: PDB Molecule:dna polymerase iii subunit psi;
PDBTitle: crystal structure of the e. coli clamp loader bound to2 primer-template dna and psi peptide
Phyre2
| 4 |
|
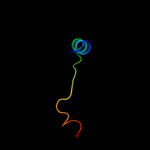
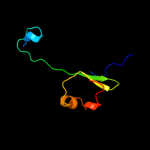
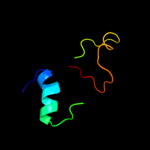
PDB 3gli chain O
Region: 2 - 28
Aligned: 27
Modelled: 27
Confidence: 98.4%
Identity: 100%
PDB header:transferase/dna
Chain: O: PDB Molecule:dna polymerase iii subunit psi;
PDBTitle: crystal structure of the e. coli clamp loader bound to2 primer-template dna and psi peptide
Phyre2
| 5 |
|
PDB 1ui0 chain A
Region: 34 - 72
Aligned: 39
Modelled: 39
Confidence: 24.1%
Identity: 18%
Fold: Uracil-DNA glycosylase-like
Superfamily: Uracil-DNA glycosylase-like
Family: Mug-like
Phyre2
| 6 |
|
PDB 1vk2 chain A
Region: 34 - 72
Aligned: 39
Modelled: 39
Confidence: 15.4%
Identity: 21%
Fold: Uracil-DNA glycosylase-like
Superfamily: Uracil-DNA glycosylase-like
Family: Mug-like
Phyre2
| 7 |
|
PDB 2kbo chain A
Region: 43 - 72
Aligned: 30
Modelled: 30
Confidence: 15.1%
Identity: 17%
PDB header:hydrolase
Chain: A: PDB Molecule:dna dc->du-editing enzyme apobec-3g;
PDBTitle: structure, interaction, and real-time monitoring of the2 enzymatic reaction of wild type apobec3g
Phyre2
| 8 |
|
PDB 2br6 chain A
Region: 12 - 71
Aligned: 60
Modelled: 60
Confidence: 14.7%
Identity: 17%
PDB header:hydrolase
Chain: A: PDB Molecule:aiia-like protein;
PDBTitle: crystal structure of quorum-quenching n-acyl homoserine2 lactone lactonase
Phyre2
| 9 |
|
PDB 3mk4 chain A
Region: 108 - 125
Aligned: 18
Modelled: 18
Confidence: 12.3%
Identity: 17%
PDB header:protein transport
Chain: A: PDB Molecule:peroxisomal biogenesis factor 3;
PDBTitle: x-ray structure of human pex3 in complex with a pex19 derived peptide
Phyre2
| 10 |
|
PDB 1taf chain A
Region: 6 - 18
Aligned: 13
Modelled: 13
Confidence: 11.4%
Identity: 15%
Fold: Histone-fold
Superfamily: Histone-fold
Family: TBP-associated factors, TAFs
Phyre2
| 11 |
|
PDB 2g80 chain C
Region: 51 - 111
Aligned: 61
Modelled: 61
Confidence: 10.9%
Identity: 13%
PDB header:hydrolase
Chain: C: PDB Molecule:protein utr4;
PDBTitle: crystal structure of utr4 protein (unknown transcript 4 protein)2 (yel038w) from saccharomyces cerevisiae at 2.28 a resolution
Phyre2
| 12 |
|
PDB 2j8p chain A
Region: 67 - 82
Aligned: 16
Modelled: 16
Confidence: 7.7%
Identity: 69%
PDB header:nuclear protein
Chain: A: PDB Molecule:cleavage stimulation factor 64 kda subunit;
PDBTitle: nmr structure of c-terminal domain of human cstf-64
Phyre2
| 13 |
|
PDB 1cmx chain A
Region: 46 - 89
Aligned: 44
Modelled: 44
Confidence: 7.0%
Identity: 23%
Fold: Cysteine proteinases
Superfamily: Cysteine proteinases
Family: Ubiquitin carboxyl-terminal hydrolase UCH-L
Phyre2
| 14 |
|
PDB 2etl chain A domain 1
Region: 46 - 89
Aligned: 44
Modelled: 44
Confidence: 6.3%
Identity: 25%
Fold: Cysteine proteinases
Superfamily: Cysteine proteinases
Family: Ubiquitin carboxyl-terminal hydrolase UCH-L
Phyre2
| 15 |
|
PDB 3o8w chain A
Region: 13 - 64
Aligned: 50
Modelled: 52
Confidence: 6.0%
Identity: 16%
PDB header:signaling protein
Chain: A: PDB Molecule:nitrogen regulatory protein p-ii (glnb-1);
PDBTitle: archaeoglobus fulgidus glnk1
Phyre2
| 16 |
|
PDB 2k29 chain A
Region: 54 - 76
Aligned: 23
Modelled: 23
Confidence: 5.9%
Identity: 30%
PDB header:transcription
Chain: A: PDB Molecule:antitoxin relb;
PDBTitle: structure of the dbd domain of e. coli antitoxin relb
Phyre2
| 17 |
|
PDB 2p5x chain B
Region: 2 - 15
Aligned: 14
Modelled: 14
Confidence: 5.6%
Identity: 29%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:n-acetylserotonin o-methyltransferase-like protein;
PDBTitle: crystal structure of maf domain of human n-acetylserotonin o-2 methyltransferase-like protein
Phyre2