| 1 |
|
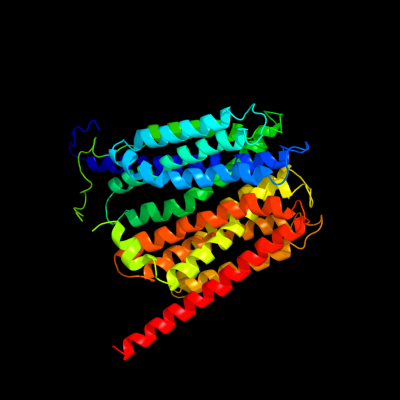
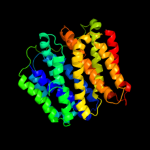
PDB 1pw4 chain A
Region: 4 - 453
Aligned: 422
Modelled: 422
Confidence: 100.0%
Identity: 13%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 2 |
|
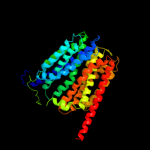
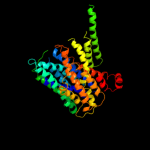
PDB 1pv7 chain A
Region: 21 - 457
Aligned: 411
Modelled: 411
Confidence: 100.0%
Identity: 12%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 3 |
|
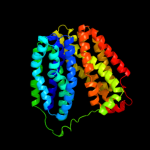
PDB 3o7p chain A
Region: 26 - 443
Aligned: 404
Modelled: 406
Confidence: 100.0%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 4 |
|
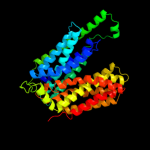
PDB 2gfp chain A
Region: 25 - 432
Aligned: 368
Modelled: 373
Confidence: 100.0%
Identity: 14%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 5 |
|
PDB 2xut chain C
Region: 27 - 439
Aligned: 412
Modelled: 413
Confidence: 100.0%
Identity: 12%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 6 |
|
PDB 3qnq chain D
Region: 406 - 458
Aligned: 53
Modelled: 53
Confidence: 43.6%
Identity: 15%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 7 |
|
PDB 1mv4 chain B
Region: 445 - 459
Aligned: 15
Modelled: 15
Confidence: 25.6%
Identity: 33%
PDB header:de novo protein
Chain: B: PDB Molecule:tropomyosin 1 alpha chain;
PDBTitle: tm9a251-284: a peptide model of the c-terminus of a rat2 striated alpha tropomyosin
Phyre2
| 8 |
|
PDB 2g9p chain A
Region: 77 - 90
Aligned: 14
Modelled: 14
Confidence: 13.6%
Identity: 29%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:antimicrobial peptide latarcin 2a;
PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
Phyre2
| 9 |
|
PDB 1cwp chain A
Region: 439 - 456
Aligned: 18
Modelled: 18
Confidence: 12.7%
Identity: 33%
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily: Positive stranded ssRNA viruses
Family: Bromoviridae-like VP
Phyre2
| 10 |
|
PDB 1cwp chain B
Region: 439 - 456
Aligned: 18
Modelled: 18
Confidence: 11.3%
Identity: 33%
PDB header:virus/rna
Chain: B: PDB Molecule:coat protein;
PDBTitle: structures of the native and swollen forms of cowpea2 chlorotic mottle virus determined by x-ray crystallography3 and cryo-electron microscopy
Phyre2
| 11 |
|
PDB 1yew chain C
Region: 418 - 456
Aligned: 39
Modelled: 39
Confidence: 8.3%
Identity: 10%
PDB header:oxidoreductase, membrane protein
Chain: C: PDB Molecule:particulate methane monooxygenase subunit c2;
PDBTitle: crystal structure of particulate methane monooxygenase
Phyre2
| 12 |
|
PDB 2qfa chain C
Region: 448 - 457
Aligned: 10
Modelled: 10
Confidence: 8.0%
Identity: 20%
PDB header:cell cycle/cell cycle/cell cycle
Chain: C: PDB Molecule:inner centromere protein;
PDBTitle: crystal structure of a survivin-borealin-incenp core complex
Phyre2
| 13 |
|
PDB 1by0 chain A
Region: 445 - 455
Aligned: 11
Modelled: 11
Confidence: 7.9%
Identity: 27%
PDB header:rna binding protein
Chain: A: PDB Molecule:protein (hepatitis delta antigen);
PDBTitle: n-terminal leucine-repeat region of hepatitis delta antigen
Phyre2
| 14 |
|
PDB 1ofc chain X domain 1
Region: 442 - 453
Aligned: 12
Modelled: 12
Confidence: 7.6%
Identity: 17%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: Myb/SANT domain
Phyre2
| 15 |
|
PDB 3mmy chain F
Region: 439 - 454
Aligned: 16
Modelled: 16
Confidence: 6.6%
Identity: 25%
PDB header:nuclear protein
Chain: F: PDB Molecule:nuclear pore complex protein nup98;
PDBTitle: structural and functional analysis of the interaction between the2 nucleoporin nup98 and the mrna export factor rae1
Phyre2
| 16 |
|
PDB 3chx chain G
Region: 427 - 456
Aligned: 30
Modelled: 30
Confidence: 6.0%
Identity: 13%
PDB header:membrane protein
Chain: G: PDB Molecule:pmoc;
PDBTitle: crystal structure of methylosinus trichosporium ob3b2 particulate methane monooxygenase (pmmo)
Phyre2
| 17 |
|
PDB 1ymg chain A
Region: 401 - 450
Aligned: 47
Modelled: 50
Confidence: 5.3%
Identity: 11%
PDB header:membrane protein
Chain: A: PDB Molecule:lens fiber major intrinsic protein;
PDBTitle: the channel architecture of aquaporin o at 2.2 angstrom resolution
Phyre2
| 18 |
|
PDB 1ymg chain A domain 1
Region: 401 - 450
Aligned: 47
Modelled: 50
Confidence: 5.3%
Identity: 11%
Fold: Aquaporin-like
Superfamily: Aquaporin-like
Family: Aquaporin-like
Phyre2