| 1 |
|
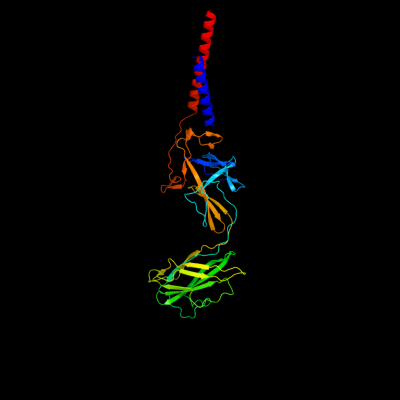
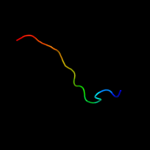
PDB 3a69 chain A
Region: 2 - 402
Aligned: 346
Modelled: 353
Confidence: 100.0%
Identity: 85%
PDB header:motor protein
Chain: A: PDB Molecule:flagellar hook protein flge;
PDBTitle: atomic model of the bacterial flagellar hook based on2 docking an x-ray derived structure and terminal two alpha-3 helices into an 7.1 angstrom resolution cryoem map
Phyre2
| 2 |
|
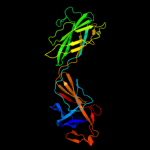
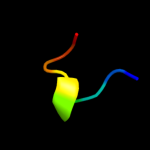
PDB 1wlg chain A
Region: 73 - 363
Aligned: 290
Modelled: 291
Confidence: 99.9%
Identity: 84%
Fold: Flagellar hook protein flgE
Superfamily: Flagellar hook protein flgE
Family: Flagellar hook protein flgE
Phyre2
| 3 |
|
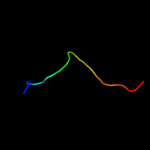
PDB 2d4y chain A
Region: 96 - 196
Aligned: 95
Modelled: 101
Confidence: 97.0%
Identity: 11%
PDB header:structural protein
Chain: A: PDB Molecule:flagellar hook-associated protein 1;
PDBTitle: crystal structure of a 49k fragment of hap1 (flgk)
Phyre2
| 4 |
|
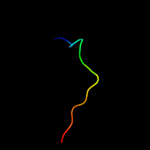
PDB 1ucu chain A
Region: 365 - 401
Aligned: 37
Modelled: 37
Confidence: 92.7%
Identity: 24%
Fold: Phase 1 flagellin
Superfamily: Phase 1 flagellin
Family: Phase 1 flagellin
Phyre2
| 5 |
|
PDB 3k8v chain B
Region: 365 - 384
Aligned: 20
Modelled: 20
Confidence: 53.8%
Identity: 30%
PDB header:structural protein
Chain: B: PDB Molecule:flagellin homolog;
PDBTitle: crysatl structure of a bacterial cell-surface flagellin n20c20
Phyre2
| 6 |
|
PDB 1ory chain B
Region: 365 - 401
Aligned: 37
Modelled: 37
Confidence: 29.7%
Identity: 22%
PDB header:chaperone
Chain: B: PDB Molecule:flagellin;
PDBTitle: flagellar export chaperone in complex with its cognate binding partner
Phyre2
| 7 |
|
PDB 1lvo chain A
Region: 23 - 37
Aligned: 15
Modelled: 15
Confidence: 13.3%
Identity: 27%
Fold: Trypsin-like serine proteases
Superfamily: Trypsin-like serine proteases
Family: Viral cysteine protease of trypsin fold
Phyre2
| 8 |
|
PDB 2q6f chain B
Region: 23 - 37
Aligned: 15
Modelled: 15
Confidence: 12.9%
Identity: 33%
PDB header:hydrolase
Chain: B: PDB Molecule:infectious bronchitis virus (ibv) main protease;
PDBTitle: crystal structure of infectious bronchitis virus (ibv) main2 protease in complex with a michael acceptor inhibitor n3
Phyre2
| 9 |
|
PDB 1nrj chain A
Region: 24 - 32
Aligned: 9
Modelled: 9
Confidence: 12.6%
Identity: 56%
Fold: Profilin-like
Superfamily: SNARE-like
Family: SRP alpha N-terminal domain-like
Phyre2
| 10 |
|
PDB 1p9s chain A
Region: 23 - 37
Aligned: 15
Modelled: 15
Confidence: 11.4%
Identity: 20%
Fold: Trypsin-like serine proteases
Superfamily: Trypsin-like serine proteases
Family: Viral cysteine protease of trypsin fold
Phyre2
| 11 |
|
PDB 3d23 chain A
Region: 23 - 37
Aligned: 15
Modelled: 15
Confidence: 9.9%
Identity: 20%
PDB header:hydrolase
Chain: A: PDB Molecule:3c-like proteinase;
PDBTitle: main protease of hcov-hku1
Phyre2
| 12 |
|
PDB 2duc chain A domain 1
Region: 23 - 37
Aligned: 15
Modelled: 15
Confidence: 9.4%
Identity: 20%
Fold: Trypsin-like serine proteases
Superfamily: Trypsin-like serine proteases
Family: Viral cysteine protease of trypsin fold
Phyre2
| 13 |
|
PDB 2npt chain A domain 1
Region: 365 - 377
Aligned: 13
Modelled: 13
Confidence: 8.9%
Identity: 23%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: CAD & PB1 domains
Family: PB1 domain
Phyre2
| 14 |
|
PDB 2gom chain A domain 1
Region: 371 - 382
Aligned: 12
Modelled: 12
Confidence: 6.6%
Identity: 17%
Fold: Spectrin repeat-like
Superfamily: Efb C-domain-like
Family: Efb C-domain-like
Phyre2
| 15 |
|
PDB 1q8h chain A
Region: 369 - 381
Aligned: 13
Modelled: 13
Confidence: 6.4%
Identity: 38%
PDB header:metal binding protein
Chain: A: PDB Molecule:osteocalcin;
PDBTitle: crystal structure of porcine osteocalcin
Phyre2
| 16 |
|
PDB 1q8h chain A
Region: 369 - 381
Aligned: 13
Modelled: 13
Confidence: 6.4%
Identity: 38%
Fold: GLA-domain
Superfamily: GLA-domain
Family: GLA-domain
Phyre2
| 17 |
|
PDB 3j0g chain O
Region: 26 - 32
Aligned: 7
Modelled: 7
Confidence: 6.3%
Identity: 29%
PDB header:virus
Chain: O: PDB Molecule:e3 protein;
PDBTitle: homology model of e3 protein of venezuelan equine encephalitis virus2 tc-83 strain fitted with a cryo-em map
Phyre2
| 18 |
|
PDB 1q3m chain A
Region: 369 - 381
Aligned: 13
Modelled: 13
Confidence: 6.2%
Identity: 38%
Fold: GLA-domain
Superfamily: GLA-domain
Family: GLA-domain
Phyre2
| 19 |
|
PDB 3ikm chain D
Region: 1 - 17
Aligned: 17
Modelled: 17
Confidence: 6.1%
Identity: 35%
PDB header:transferase
Chain: D: PDB Molecule:dna polymerase subunit gamma-1;
PDBTitle: crystal structure of human mitochondrial dna polymerase2 holoenzyme
Phyre2
| 20 |
|
PDB 2dld chain A domain 2
Region: 22 - 34
Aligned: 13
Modelled: 13
Confidence: 6.0%
Identity: 0%
Fold: Flavodoxin-like
Superfamily: Formate/glycerate dehydrogenase catalytic domain-like
Family: Formate/glycerate dehydrogenases, substrate-binding domain
Phyre2
| 21 |
|
| 22 |
|