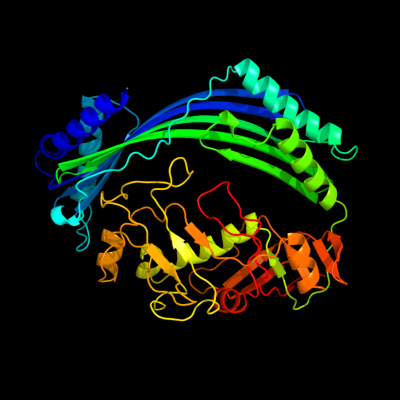
| 1 | c3qtdC_
|
|
 |
100.0 |
50 |
PDB header:gene regulation
Chain: C: PDB Molecule:pmba protein;
PDBTitle: crystal structure of putative modulator of gyrase (pmba) from2 pseudomonas aeruginosa pao1
|
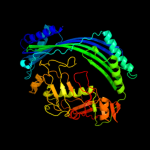
| 2 | d1vpba_
|
|
 |
100.0 |
28 |
Fold:Putative modulator of DNA gyrase, PmbA/TldD
Superfamily:Putative modulator of DNA gyrase, PmbA/TldD
Family:Putative modulator of DNA gyrase, PmbA/TldD |
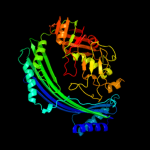
| 3 | d1vl4a_
|
|
 |
100.0 |
28 |
Fold:Putative modulator of DNA gyrase, PmbA/TldD
Superfamily:Putative modulator of DNA gyrase, PmbA/TldD
Family:Putative modulator of DNA gyrase, PmbA/TldD |
| 4 | c3onrI_
|
|
 |
25.1 |
12 |
PDB header:metal binding protein
Chain: I: PDB Molecule:protein transport protein sece2;
PDBTitle: crystal structure of the calcium chelating immunodominant antigen,2 calcium dodecin (rv0379),from mycobacterium tuberculosis with a novel3 calcium-binding site
|
| 5 | d1o12a1
|
|
 |
24.5 |
29 |
Fold:Composite domain of metallo-dependent hydrolases
Superfamily:Composite domain of metallo-dependent hydrolases
Family:N-acetylglucosamine-6-phosphate deacetylase, NagA |
| 6 | d2ux9a1
|
|
 |
23.2 |
16 |
Fold:Dodecin subunit-like
Superfamily:Dodecin-like
Family:Dodecin-like |
| 7 | c2vxaL_
|
|
 |
23.0 |
16 |
PDB header:flavoprotein
Chain: L: PDB Molecule:dodecin;
PDBTitle: h.halophila dodecin in complex with riboflavin
|
| 8 | c3oqtP_
|
|
 |
22.6 |
14 |
PDB header:flavoprotein
Chain: P: PDB Molecule:rv1498a protein;
PDBTitle: crystal structure of rv1498a protein from mycobacterium tuberculosis
|
| 9 | c3gknA_
|
|
 |
11.9 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:bacterioferritin comigratory protein;
PDBTitle: insights into the alkyl peroxide reduction activity of xanthomonas2 campestris bacterioferritin comigratory protein from the trapped3 intermediate/ligand complex structures
|
| 10 | d1xaca_
|
|
 |
11.5 |
18 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 11 | d2pjua1
|
|
 |
9.2 |
21 |
Fold:Chelatase-like
Superfamily:PrpR receptor domain-like
Family:PrpR receptor domain-like |
| 12 | c2d1cB_
|
|
 |
8.9 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: crystal structure of tt0538 protein from thermus thermophilus hb8
|
| 13 | d1pb1a_
|
|
 |
8.0 |
15 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 14 | c3ixrA_
|
|
 |
7.6 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:bacterioferritin comigratory protein;
PDBTitle: crystal structure of xylella fastidiosa prxq c47s mutant
|
| 15 | c2iv0A_
|
|
 |
7.2 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: thermal stability of isocitrate dehydrogenase from2 archaeoglobus fulgidus studied by crystal structure3 analysis and engineering of chimers
|
| 16 | d1w0da_
|
|
 |
6.8 |
19 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 17 | d1hqsa_
|
|
 |
6.7 |
12 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 18 | d1jjga_
|
|
 |
6.7 |
38 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 19 | d1n8ja_
|
|
 |
6.4 |
23 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 20 | c3blxM_
|
|
 |
6.4 |
15 |
PDB header:oxidoreductase
Chain: M: PDB Molecule:isocitrate dehydrogenase [nad] subunit 1;
PDBTitle: yeast isocitrate dehydrogenase (apo form)
|
| 21 | c3f7xA_ |
|
not modelled |
6.2 |
29 |
PDB header:unknown function
Chain: A: PDB Molecule:putative polyketide cyclase;
PDBTitle: crystal structure of a putative polyketide cyclase (pp0894) from2 pseudomonas putida kt2440 at 1.24 a resolution
|
| 22 | c2kilA_ |
|
not modelled |
6.2 |
6 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: nmr structure of the h103g mutant so2144 h-nox domain from2 shewanella oneidensis in the fe(ii)co ligation state
|
| 23 | c3r8wC_ |
|
not modelled |
6.1 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:3-isopropylmalate dehydrogenase 2, chloroplastic;
PDBTitle: structure of 3-isopropylmalate dehydrogenase isoform 2 from2 arabidopsis thaliana at 2.2 angstrom resolution
|
| 24 | c3e8vA_ |
|
not modelled |
6.1 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:possible transglutaminase-family protein;
PDBTitle: crystal structure of a possible transglutaminase-family2 protein proteolytic fragment from bacteroides fragilis
|
| 25 | d1a05a_ |
|
not modelled |
6.1 |
8 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 26 | c1bknA_ |
|
not modelled |
5.9 |
15 |
PDB header:dna repair
Chain: A: PDB Molecule:mutl;
PDBTitle: crystal structure of an n-terminal 40kd fragment of e. coli2 dna mismatch repair protein mutl
|
| 27 | d1ydua1 |
|
not modelled |
5.9 |
9 |
Fold:At5g01610-like
Superfamily:At5g01610-like
Family:At5g01610-like |
| 28 | d1cm7a_ |
|
not modelled |
5.9 |
12 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 29 | c3mhsE_ |
|
not modelled |
5.9 |
15 |
PDB header:hydrolase/transcription regulator/protei
Chain: E: PDB Molecule:saga-associated factor 73;
PDBTitle: structure of the saga ubp8/sgf11/sus1/sgf73 dub module bound to2 ubiquitin aldehyde
|
| 30 | d3ckma1 |
|
not modelled |
5.8 |
13 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 31 | d1luza_ |
|
not modelled |
5.8 |
22 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 32 | d1ylea1 |
|
not modelled |
5.7 |
6 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:AstA-like |
| 33 | c1tyoA_ |
|
not modelled |
5.7 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: isocitrate dehydrogenase from the hyperthermophile aeropyrum pernix in2 complex with etheno-nadp
|
| 34 | d1wpwa_ |
|
not modelled |
5.5 |
23 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 35 | d1b25a1 |
|
not modelled |
5.5 |
15 |
Fold:Aldehyde ferredoxin oxidoreductase, C-terminal domains
Superfamily:Aldehyde ferredoxin oxidoreductase, C-terminal domains
Family:Aldehyde ferredoxin oxidoreductase, C-terminal domains |
| 36 | c2kk4A_ |
|
not modelled |
5.5 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein af_2094;
PDBTitle: solution nmr structure of protein af2094 from archaeoglobus2 fulgidus. northeast structural genomics consotium (nesg)3 target gt2
|
| 37 | c2op8A_ |
|
not modelled |
5.5 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:probable tautomerase ywhb;
PDBTitle: crystal structure of ywhb- homologue of 4-oxalocrotonate tautomerase
|
| 38 | d1m1ha1 |
|
not modelled |
5.4 |
46 |
Fold:N-utilization substance G protein NusG, insert domain
Superfamily:N-utilization substance G protein NusG, insert domain
Family:N-utilization substance G protein NusG, insert domain |
| 39 | c1htrP_ |
|
not modelled |
5.4 |
29 |
PDB header:aspartyl protease
Chain: P: PDB Molecule:progastricsin (pro segment);
PDBTitle: crystal and molecular structures of human progastricsin at 1.622 angstroms resolution
|
| 40 | d1dl6a_ |
|
not modelled |
5.3 |
17 |
Fold:Rubredoxin-like
Superfamily:Zinc beta-ribbon
Family:Transcriptional factor domain |
| 41 | d1ka1a_ |
|
not modelled |
5.3 |
18 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 42 | d2q0ia1 |
|
not modelled |
5.2 |
22 |
Fold:Metallo-hydrolase/oxidoreductase
Superfamily:Metallo-hydrolase/oxidoreductase
Family:PqsE-like |
| 43 | d1cnza_ |
|
not modelled |
5.2 |
12 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 44 | d1yrra2 |
|
not modelled |
5.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:N-acetylglucosamine-6-phosphate deacetylase, NagA, catalytic domain |
| 45 | c3dbaB_ |
|
not modelled |
5.2 |
10 |
PDB header:hydrolase
Chain: B: PDB Molecule:cone cgmp-specific 3',5'-cyclic phosphodiesterase subunit
PDBTitle: crystal structure of the cgmp-bound gaf a domain from the2 photoreceptor phosphodiesterase 6c
|
| 46 | d2h6ca2 |
|
not modelled |
5.1 |
17 |
Fold:Double-stranded beta-helix
Superfamily:cAMP-binding domain-like
Family:cAMP-binding domain |
| 47 | d1aora1 |
|
not modelled |
5.1 |
8 |
Fold:Aldehyde ferredoxin oxidoreductase, C-terminal domains
Superfamily:Aldehyde ferredoxin oxidoreductase, C-terminal domains
Family:Aldehyde ferredoxin oxidoreductase, C-terminal domains |
| 48 | d1g2ua_ |
|
not modelled |
5.1 |
12 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 49 | c3hz4A_ |
|
not modelled |
5.1 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin;
PDBTitle: crystal structure of thioredoxin from methanosarcina mazei
|
| 50 | d1u55a_ |
|
not modelled |
5.1 |
6 |
Fold:Ligand-binding domain in the NO signalling and Golgi transport
Superfamily:Ligand-binding domain in the NO signalling and Golgi transport
Family:H-NOX domain |
| 51 | c2ideE_ |
|
not modelled |
5.1 |
25 |
PDB header:biosynthetic protein
Chain: E: PDB Molecule:molybdenum cofactor biosynthesis protein c;
PDBTitle: crystal structure of the molybdenum cofactor biosynthesis protein c2 (ttha1789) from thermus theromophilus hb8
|