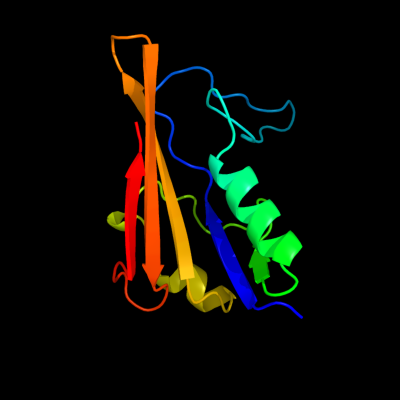
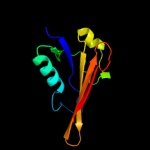
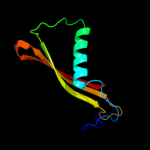
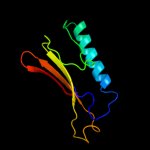
1 d2hqsa2
98.3
21
Fold: Anticodon-binding domain-likeSuperfamily: TolB, N-terminal domainFamily: TolB, N-terminal domain2 d2iqia1
96.4
15
Fold: Anticodon-binding domain-likeSuperfamily: XCC0632-likeFamily: XCC0632-like3 c3ly8A_
87.5
13
PDB header: signaling proteinChain: A: PDB Molecule: transcriptional activator cadc;PDBTitle: crystal structure of mutant d471e of the periplasmic domain of cadc
4 d1wuea2
41.0
18
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like5 c2w8bB_
32.6
20
PDB header: protein transport/membrane proteinChain: B: PDB Molecule: protein tolb;PDBTitle: crystal structure of processed tolb in complex with pal
6 d2czra1
21.3
23
Fold: Restriction endonuclease-likeSuperfamily: TBP-interacting protein-likeFamily: TBP-interacting protein-like7 d2gdqa2
20.5
13
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like8 d2mnra2
20.5
10
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like9 d1sjda2
19.7
15
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like10 c3lo7A_
18.6
40
PDB header: transferaseChain: A: PDB Molecule: penicillin-binding protein a;PDBTitle: crystal structure of pbpa from mycobacterium tuberculosis
11 d1wufa2
17.8
19
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like12 d1jpma2
17.8
11
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like13 d1jpdx2
17.3
19
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like14 c3ue3A_
16.9
47
PDB header: transferaseChain: A: PDB Molecule: septum formation, penicillin binding protein 3,PDBTitle: crystal structure of acinetobacter baumanni pbp3
15 d1jdfa2
16.6
14
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like16 d1r0ma2
16.5
15
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like17 d2gl5a2
16.3
19
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like18 d2chra2
16.2
15
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like19 d1nu5a2
15.0
19
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like20 d1rp5a4
14.5
40
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase21 d1vqqa3
not modelled
14.4
40
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase22 d1k25a4
not modelled
14.2
40
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase23 d1muca2
not modelled
12.4
15
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like24 c3equB_
not modelled
11.0
53
PDB header: biosynthetic proteinChain: B: PDB Molecule: penicillin-binding protein 2;PDBTitle: crystal structure of penicillin-binding protein 2 from neisseria2 gonorrhoeae
25 d1bqga2
not modelled
10.2
13
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like26 c3pbqA_
not modelled
9.6
47
PDB header: hydrolase/antibioticChain: A: PDB Molecule: penicillin-binding protein 3;PDBTitle: crystal structure of pbp3 complexed with imipenem
27 c3mbrX_
not modelled
9.3
25
PDB header: transferaseChain: X: PDB Molecule: glutamine cyclotransferase;PDBTitle: crystal structure of the glutaminyl cyclase from xanthomonas2 campestris
28 d1pyya4
not modelled
9.2
40
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase29 c1mwuA_
not modelled
9.0
40
PDB header: biosynthetic proteinChain: A: PDB Molecule: penicillin-binding protein 2a;PDBTitle: structure of methicillin acyl-penicillin binding protein 2a2 from methicillin resistant staphylococcus aureus strain3 27r at 2.60 a resolution.
30 d2c5wb1
not modelled
8.7
29
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase31 d1tw3a1
not modelled
8.2
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Plant O-methyltransferase, N-terminal domain32 c2xzmE_
not modelled
8.1
17
PDB header: ribosomeChain: E: PDB Molecule: ribosomal protein s5 containing protein;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
33 c3nqyA_
not modelled
8.0
42
PDB header: hydrolaseChain: A: PDB Molecule: secreted metalloprotease mcp02;PDBTitle: crystal structure of the autoprocessed complex of vibriolysin mcp-022 with a single point mutation e346a
34 c3oc2A_
not modelled
7.8
47
PDB header: penicillin-binding proteinChain: A: PDB Molecule: penicillin-binding protein 3;PDBTitle: crystal structure of penicillin-binding protein 3 from pseudomonas2 aeruginosa
35 c2wadB_
not modelled
6.6
33
PDB header: peptide binding proteinChain: B: PDB Molecule: penicillin-binding protein 2b;PDBTitle: penicillin-binding protein 2b (pbp-2b) from streptococcus2 pneumoniae (strain 5204)
36 d2bg1a1
not modelled
6.2
36
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase37 d1svda1
not modelled
6.2
20
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain38 c1x37A_
not modelled
6.0
21
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent protease la 1;PDBTitle: structure of bacillus subtilis lon protease ssd domain
39 c2kbxA_
not modelled
5.8
11
PDB header: cell adhesionChain: A: PDB Molecule: integrin-linked protein kinase;PDBTitle: solution structure of ilk-pinch complex
40 c3cb3B_
not modelled
5.7
12
PDB header: isomeraseChain: B: PDB Molecule: mandelate racemase/muconate lactonizing enzyme;PDBTitle: crystal structure of l-talarate dehydratase from polaromonas sp. js6662 complexed with mg and l-glucarate
41 d1bwva1
not modelled
5.6
17
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain42 c1qmfA_
not modelled
5.5
40
PDB header: peptidoglycan synthesisChain: A: PDB Molecule: penicillin-binding protein 2x;PDBTitle: penicillin-binding protein 2x (pbp-2x) acyl-enzyme complex
43 d2hc5a1
not modelled
5.5
29
Fold: FlaG-likeSuperfamily: FlaG-likeFamily: FlaG-like44 d1qzza1
not modelled
5.5
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Plant O-methyltransferase, N-terminal domain45 d2olua2
not modelled
5.3
14
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase46 c3onrI_
not modelled
5.2
14
PDB header: metal binding proteinChain: I: PDB Molecule: protein transport protein sece2;PDBTitle: crystal structure of the calcium chelating immunodominant antigen,2 calcium dodecin (rv0379),from mycobacterium tuberculosis with a novel3 calcium-binding site