1 c3fggA_
32.7
46
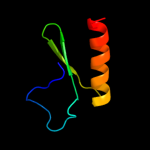
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein bce2196;PDBTitle: crystal structure of putative ecf-type sigma factor negative effector2 from bacillus cereus
2 c1nw1A_
22.4
23
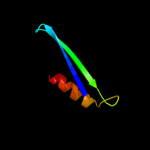
PDB header: transferaseChain: A: PDB Molecule: choline kinase (49.2 kd);PDBTitle: crystal structure of choline kinase
3 d1nw1a_
22.4
23
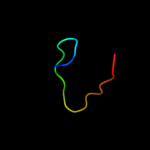
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: Choline kinase4 d1v5ma_
21.7
26
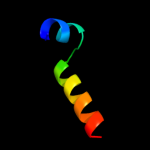
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Pleckstrin-homology domain (PH domain)5 c3mesB_
17.9
16
PDB header: transferaseChain: B: PDB Molecule: choline kinase;PDBTitle: crystal structure of choline kinase from cryptosporidium2 parvum iowa ii, cgd3_2030
6 d1bbpa_
14.0
23
Fold: LipocalinsSuperfamily: LipocalinsFamily: Retinol binding protein-like7 c2hzqA_
13.8
17
PDB header: transport proteinChain: A: PDB Molecule: apolipoprotein d;PDBTitle: crystal structure of human apolipoprotein d (apod) in2 complex with progesterone
8 d2ox7a1
13.7
33
Fold: YopX-likeSuperfamily: YopX-likeFamily: YopX-like9 c3ha4C_
13.4
24
PDB header: unknown functionChain: C: PDB Molecule: mix1;PDBTitle: crystal structure of the type one membrane protein mix1 from2 leishmania
10 d1gkab_
12.7
13
Fold: LipocalinsSuperfamily: LipocalinsFamily: Retinol binding protein-like11 d1axia_
12.3
27
Fold: 4-helical cytokinesSuperfamily: 4-helical cytokinesFamily: Long-chain cytokines12 d2b7ea1
11.2
43
Fold: Another 3-helical bundleSuperfamily: FF domainFamily: FF domain13 c1zxjB_
10.7
21
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein mg377 homolog;PDBTitle: crystal structure of the hypthetical mycoplasma protein,2 mpn555
14 c1h4tD_
10.4
25
PDB header: aminoacyl-trna synthetaseChain: D: PDB Molecule: prolyl-trna synthetase;PDBTitle: prolyl-trna synthetase from thermus thermophilus complexed2 with l-proline
15 d2p84a1
10.0
31
Fold: YopX-likeSuperfamily: YopX-likeFamily: YopX-like16 c2qyzA_
9.9
38
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the uncharacterized protein ctc02137 from2 clostridium tetani e88
17 d1osce_
9.0
13
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)18 d1pm1x_
8.8
13
Fold: LipocalinsSuperfamily: LipocalinsFamily: Retinol binding protein-like19 c3c8uA_
8.7
14
PDB header: transferaseChain: A: PDB Molecule: fructokinase;PDBTitle: crystal structure of putative fructose transport system kinase2 (yp_612366.1) from silicibacter sp. tm1040 at 1.95 a resolution
20 d1whra_
8.0
14
Fold: IF3-likeSuperfamily: R3H domainFamily: R3H domain21 c1wqdA_
not modelled
7.9
46
PDB header: toxinChain: A: PDB Molecule: omtx2;PDBTitle: an unusual fold for potassium channel blockers: nmr2 structure of three toxins from the scorpion opisthacanthus3 madagascariensis
22 c3fwlA_
not modelled
7.9
21
PDB header: transferase, hydrolaseChain: A: PDB Molecule: penicillin-binding protein 1b;PDBTitle: crystal structure of the full-length transglycosylase pbp1b2 from escherichia coli
23 c3lx6B_
not modelled
7.7
21
PDB header: transferaseChain: B: PDB Molecule: cytosine-specific methyltransferase;PDBTitle: structure of probable cytosine-specific methyltransferase from2 shigella flexneri
24 d1hgua_
not modelled
7.4
17
Fold: 4-helical cytokinesSuperfamily: 4-helical cytokinesFamily: Long-chain cytokines25 c1wqcA_
not modelled
7.3
46
PDB header: toxinChain: A: PDB Molecule: omtx1;PDBTitle: an unusual fold for potassium channel blockers : nmr2 structure of three toxins from the scorpion opisthacanthus3 madagascariensis
26 d1v65a_
not modelled
7.2
25
Fold: KRAB domain (Kruppel-associated box)Superfamily: KRAB domain (Kruppel-associated box)Family: KRAB domain (Kruppel-associated box)27 d1mg4a_
not modelled
7.2
40
Fold: beta-Grasp (ubiquitin-like)Superfamily: Doublecortin (DC)Family: Doublecortin (DC)28 c2l23A_
not modelled
7.0
58
PDB header: transcriptionChain: A: PDB Molecule: mediator of rna polymerase ii transcription subunit 25;PDBTitle: nmr structure of the acid (activator interacting domain) of the human2 mediator med25 protein
29 c3rfrI_
not modelled
6.9
13
PDB header: oxidoreductaseChain: I: PDB Molecule: pmob;PDBTitle: crystal structure of particulate methane monooxygenase (pmmo) from2 methylocystis sp. strain m
30 d1m8na_
not modelled
6.6
46
Fold: Single-stranded left-handed beta-helixSuperfamily: An insect antifreeze proteinFamily: An insect antifreeze protein31 d1clvi_
not modelled
6.5
71
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Plant inhibitors of proteinases and amylasesFamily: Plant inhibitors of proteinases and amylases32 c3rgbA_
not modelled
6.3
19
PDB header: oxidoreductaseChain: A: PDB Molecule: methane monooxygenase subunit b2;PDBTitle: crystal structure of particulate methane monooxygenase from2 methylococcus capsulatus (bath)
33 c1yewI_
not modelled
6.3
19
PDB header: oxidoreductase, membrane proteinChain: I: PDB Molecule: particulate methane monooxygenase, b subunit;PDBTitle: crystal structure of particulate methane monooxygenase
34 c2qrvA_
not modelled
6.3
22
PDB header: transferase/transferase regulatorChain: A: PDB Molecule: dna (cytosine-5)-methyltransferase 3a;PDBTitle: structure of dnmt3a-dnmt3l c-terminal domain complex
35 c3dwlF_
not modelled
6.3
20
PDB header: structural proteinChain: F: PDB Molecule: actin-related protein 2/3 complex subunit 4;PDBTitle: crystal structure of fission yeast arp2/3 complex lacking the arp22 subunit
36 d1ewna_
not modelled
6.2
27
Fold: FMT C-terminal domain-likeSuperfamily: FMT C-terminal domain-likeFamily: 3-methyladenine DNA glycosylase (AAG, ANPG, MPG)37 c3chxE_
not modelled
6.0
12
PDB header: membrane proteinChain: E: PDB Molecule: pmob;PDBTitle: crystal structure of methylosinus trichosporium ob3b2 particulate methane monooxygenase (pmmo)
38 c2qrvC_
not modelled
5.9
20
PDB header: transferase/transferase regulatorChain: C: PDB Molecule: dna (cytosine-5)-methyltransferase 3-like;PDBTitle: structure of dnmt3a-dnmt3l c-terminal domain complex
39 d1wjpa2
not modelled
5.9
46
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H240 d1v7wa2
not modelled
5.8
13
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Glycosyltransferase family 36 N-terminal domain41 d1n0sa_
not modelled
5.8
20
Fold: LipocalinsSuperfamily: LipocalinsFamily: Retinol binding protein-like42 d1mjda_
not modelled
5.8
33
Fold: beta-Grasp (ubiquitin-like)Superfamily: Doublecortin (DC)Family: Doublecortin (DC)43 d2axla1
not modelled
5.5
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RecQ helicase DNA-binding domain-like44 d1e6ya1
not modelled
5.2
46
Fold: Methyl-coenzyme M reductase alpha and beta chain C-terminal domainSuperfamily: Methyl-coenzyme M reductase alpha and beta chain C-terminal domainFamily: Methyl-coenzyme M reductase alpha and beta chain C-terminal domain45 c3c5iD_
not modelled
5.2
16
PDB header: transferaseChain: D: PDB Molecule: choline kinase;PDBTitle: crystal structure of plasmodium knowlesi choline kinase, pkh_134520
46 c1ydiB_
not modelled
5.1
37
PDB header: cell adhesion, structural proteinChain: B: PDB Molecule: alpha-actinin 4;PDBTitle: human vinculin head domain (vh1, 1-258) in complex with2 human alpha-actinin's vinculin-binding site (residues 731-3 760)
47 c1zx3A_
not modelled
5.1
30
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ne0241;PDBTitle: structure of ne0241 protein of unknown function from nitrosomonas2 europaea