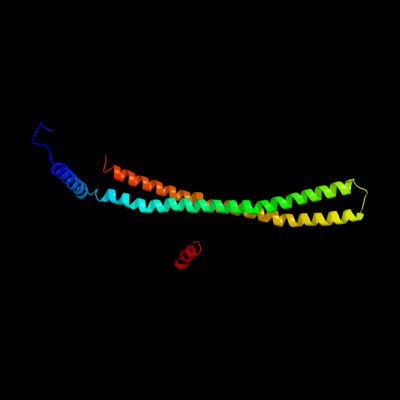
1 c1kmiZ_
100.0
100
PDB header: signaling proteinChain: Z: PDB Molecule: chemotaxis protein chez;PDBTitle: crystal structure of an e.coli chemotaxis protein, chez
2 c2pl9D_
97.1
100
PDB header: signaling protienChain: D: PDB Molecule: chemotaxis protein chez;PDBTitle: crystal structure of chey-mg(2+)-bef(3)(-) in complex with chez(c19)2 peptide solved from a p2(1)2(1)2 crystal
3 c2pl9E_
95.9
100
PDB header: signaling protienChain: E: PDB Molecule: chemotaxis protein chez;PDBTitle: crystal structure of chey-mg(2+)-bef(3)(-) in complex with chez(c19)2 peptide solved from a p2(1)2(1)2 crystal
4 c2pl9F_
95.9
100
PDB header: signaling protienChain: F: PDB Molecule: chemotaxis protein chez;PDBTitle: crystal structure of chey-mg(2+)-bef(3)(-) in complex with chez(c19)2 peptide solved from a p2(1)2(1)2 crystal
5 c2damA_
43.1
75
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: etea protein;PDBTitle: solution structure of the novel identified uba-like domain2 in the n-terminal of human etea protein
6 c1zvzA_
35.4
14
PDB header: protein bindingChain: A: PDB Molecule: vinculin;PDBTitle: vinculin head (0-258) in complex with the talin rod residue2 820-844
7 d1szia_
34.5
15
Fold: Four-helical up-and-down bundleSuperfamily: Mannose-6-phosphate receptor binding protein 1 (Tip47), C-terminal domainFamily: Mannose-6-phosphate receptor binding protein 1 (Tip47), C-terminal domain8 d2a2pa1
32.2
33
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Selenoprotein W-related9 d1rqba1
30.8
20
Fold: RuvA C-terminal domain-likeSuperfamily: post-HMGL domain-likeFamily: Conserved carboxylase domain10 c2wpqA_
27.6
7
PDB header: membrane proteinChain: A: PDB Molecule: trimeric autotransporter adhesin fragment;PDBTitle: salmonella enterica sada 479-519 fused to gcn4 adaptors (2 sadak3, in-register fusion)
11 c2vs0B_
21.7
9
PDB header: cell invasionChain: B: PDB Molecule: virulence factor esxa;PDBTitle: structural analysis of homodimeric staphylococcal aureus2 virulence factor esxa
12 c3gvmA_
18.2
13
PDB header: viral proteinChain: A: PDB Molecule: putative uncharacterized protein sag1039;PDBTitle: structure of the homodimeric wxg-100 family protein from streptococcus2 agalactiae
13 c1rr2A_
18.1
20
PDB header: transferaseChain: A: PDB Molecule: transcarboxylase 5s subunit;PDBTitle: propionibacterium shermanii transcarboxylase 5s subunit bound to 2-2 ketobutyric acid
14 d1wa8a1
15.4
15
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like15 c3hkzZ_
14.5
33
PDB header: transferaseChain: Z: PDB Molecule: dna-directed rna polymerase subunit 13;PDBTitle: the x-ray crystal structure of rna polymerase from archaea
16 c3ayhA_
14.3
13
PDB header: transcriptionChain: A: PDB Molecule: dna-directed rna polymerase iii subunit rpc9;PDBTitle: crystal structure of the c17/25 subcomplex from s. pombe rna2 polymerase iii
17 c3hkzY_
13.8
32
PDB header: transferaseChain: Y: PDB Molecule: dna-directed rna polymerase subunit 13;PDBTitle: the x-ray crystal structure of rna polymerase from archaea
18 c2waqQ_
13.6
28
PDB header: transcriptionChain: Q: PDB Molecule: dna-directed rna polymerase rpo13 subunit;PDBTitle: the complete structure of the archaeal 13-subunit dna-2 directed rna polymerase
19 d2e1fa1
12.5
10
Fold: SAM domain-likeSuperfamily: HRDC-likeFamily: HRDC domain from helicases20 c2y0sJ_
12.0
30
PDB header: transferaseChain: J: PDB Molecule: rna polymerase subunit 13;PDBTitle: crystal structure of sulfolobus shibatae rna polymerase in2 p21 space group
21 c2y0sQ_
not modelled
12.0
30
PDB header: transferaseChain: Q: PDB Molecule: rna polymerase subunit 13;PDBTitle: crystal structure of sulfolobus shibatae rna polymerase in2 p21 space group
22 c2nx9B_
not modelled
11.7
20
PDB header: lyaseChain: B: PDB Molecule: oxaloacetate decarboxylase 2, subunit alpha;PDBTitle: crystal structure of the carboxyltransferase domain of the2 oxaloacetate decarboxylase na+ pump from vibrio cholerae
23 d1wa8b1
not modelled
11.3
12
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like24 d1we1a_
not modelled
10.9
22
Fold: Heme oxygenase-likeSuperfamily: Heme oxygenase-likeFamily: Eukaryotic type heme oxygenase25 c2wb1Q_
not modelled
10.3
30
PDB header: transcriptionChain: Q: PDB Molecule: dna-directed rna polymerase rpo13 subunit;PDBTitle: the complete structure of the archaeal 13-subunit dna-2 directed rna polymerase
26 c2dnxA_
not modelled
10.1
13
PDB header: transport proteinChain: A: PDB Molecule: syntaxin-12;PDBTitle: solution structure of rsgi ruh-063, an n-terminal domain of2 syntaxin 12 from human cdna
27 c2wb1J_
not modelled
9.7
30
PDB header: transcriptionChain: J: PDB Molecule: dna-directed rna polymerase rpo13 subunit;PDBTitle: the complete structure of the archaeal 13-subunit dna-2 directed rna polymerase
28 d2fyma2
not modelled
9.6
20
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like29 c3g67A_
not modelled
9.4
12
PDB header: signaling proteinChain: A: PDB Molecule: methyl-accepting chemotaxis protein;PDBTitle: crystal structure of a soluble chemoreceptor from thermotoga2 maritima
30 d1eija_
not modelled
9.3
50
Fold: RuvA C-terminal domain-likeSuperfamily: Double-stranded DNA-binding domainFamily: Double-stranded DNA-binding domain31 c2vzdD_
not modelled
9.2
50
PDB header: cell adhesionChain: D: PDB Molecule: paxillin;PDBTitle: crystal structure of the c-terminal calponin homology2 domain of alpha parvin in complex with paxillin ld1 motif
32 c2krcA_
not modelled
9.1
13
PDB header: transcriptionChain: A: PDB Molecule: dna-directed rna polymerase subunit delta;PDBTitle: solution structure of the n-terminal domain of bacillus2 subtilis delta subunit of rna polymerase
33 c2pmzF_
not modelled
9.1
16
PDB header: translation, transferaseChain: F: PDB Molecule: dna-directed rna polymerase subunit f;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
34 c2vzdC_
not modelled
9.0
50
PDB header: cell adhesionChain: C: PDB Molecule: paxillin;PDBTitle: crystal structure of the c-terminal calponin homology2 domain of alpha parvin in complex with paxillin ld1 motif
35 d2dnaa1
not modelled
8.4
31
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain36 d1dd5a_
not modelled
8.2
14
Fold: RRF/tRNA synthetase additional domain-likeSuperfamily: Ribosome recycling factor, RRFFamily: Ribosome recycling factor, RRF37 d2akza2
not modelled
7.6
33
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like38 d1ug7a_
not modelled
7.4
19
Fold: Four-helical up-and-down bundleSuperfamily: Domain from hypothetical 2610208m17rik proteinFamily: Domain from hypothetical 2610208m17rik protein39 d2al1a2
not modelled
7.3
53
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like40 d1pdza2
not modelled
7.1
27
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like41 c3k8wA_
not modelled
7.0
17
PDB header: structural proteinChain: A: PDB Molecule: flagellin homolog;PDBTitle: crysatl structure of a bacterial cell-surface flagellin n20c45
42 c3s90B_
not modelled
6.7
14
PDB header: cell adhesionChain: B: PDB Molecule: vinculin;PDBTitle: human vinculin head domain vh1 (residues 1-252) in complex with murine2 talin (vbs33; residues 1512-1546)
43 d2dsma1
not modelled
6.6
40
Fold: YqaI-likeSuperfamily: YqaI-likeFamily: YqaI-like44 c2drnC_
not modelled
6.6
36
PDB header: transferaseChain: C: PDB Molecule: 24-residues peptide from an a-kinase anchoringPDBTitle: docking and dimerization domain (d/d) of the type ii-alpha2 regulatory subunity of protein kinase a (pka) in complex3 with a peptide from an a-kinase anchoring protein
45 d1jjcb2
not modelled
6.2
22
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: Domains B1 and B5 of PheRS-beta, PheT46 d1st6a5
not modelled
6.1
15
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: alpha-catenin/vinculin47 c2a93B_
not modelled
6.0
57
PDB header: leucine zippersChain: B: PDB Molecule: c-myc-max heterodimeric leucine zipper;PDBTitle: nmr solution structure of the c-myc-max heterodimeric2 leucine zipper, 40 structures
48 d1qf9a_
not modelled
5.9
38
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases49 d1u2ma_
not modelled
5.7
40
Fold: OmpH-likeSuperfamily: OmpH-likeFamily: OmpH-like50 c2k5cA_
not modelled
5.7
27
PDB header: metal binding proteinChain: A: PDB Molecule: uncharacterized protein pf0385;PDBTitle: nmr structure for pf0385
51 d1s94a_
not modelled
5.5
14
Fold: STAT-likeSuperfamily: t-snare proteinsFamily: t-snare proteins52 c1s94A_
not modelled
5.5
14
PDB header: endocytosis/exocytosisChain: A: PDB Molecule: s-syntaxin;PDBTitle: crystal structure of the habc domain of neuronal syntaxin from the2 squid loligo pealei
53 c3cdhB_
not modelled
5.4
19
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, marr family;PDBTitle: crystal structure of the marr family transcriptional regulator spo14532 from silicibacter pomeroyi dss-3
54 c2ch7A_
not modelled
5.2
14
PDB header: chemotaxisChain: A: PDB Molecule: methyl-accepting chemotaxis protein;PDBTitle: crystal structure of the cytoplasmic domain of a bacterial2 chemoreceptor from thermotoga maritima
55 c3tj5A_
not modelled
5.1
17
PDB header: protein binding/toxinChain: A: PDB Molecule: vinculin;PDBTitle: human vinculin head domain (vh1, residues 1-258) in complex with the2 vinculin binding site of the surface cell antigen 4 (sca4-vbs-n;3 residues 412-434) from rickettsia rickettsii