| 1 |
|
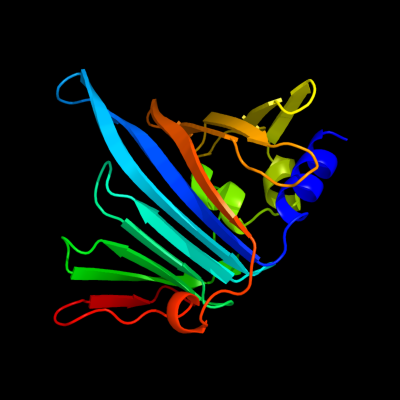
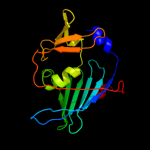
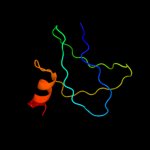
PDB 1iwl chain A
Region: 22 - 203
Aligned: 177
Modelled: 182
Confidence: 100.0%
Identity: 99%
Fold: LolA-like prokaryotic lipoproteins and lipoprotein localization factors
Superfamily: Prokaryotic lipoproteins and lipoprotein localization factors
Family: Outer-membrane lipoproteins carrier protein LolA
Phyre2
| 2 |
|
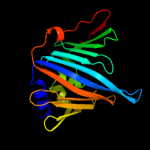
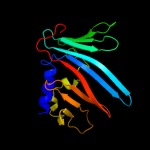
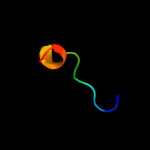
PDB 2w7q chain B
Region: 21 - 201
Aligned: 174
Modelled: 181
Confidence: 100.0%
Identity: 35%
PDB header:protein transport
Chain: B: PDB Molecule:outer-membrane lipoprotein carrier protein;
PDBTitle: structure of pseudomonas aeruginosa lola
Phyre2
| 3 |
|
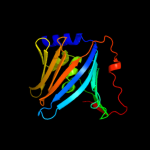
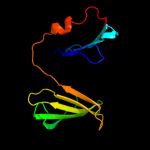
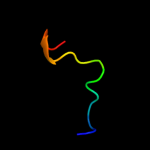
PDB 3buu chain B
Region: 17 - 189
Aligned: 170
Modelled: 173
Confidence: 99.7%
Identity: 14%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized lola superfamily protein ne2245;
PDBTitle: crystal structure of lola superfamily protein ne2245 from2 nitrosomonas europaea
Phyre2
| 4 |
|
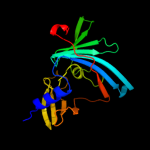
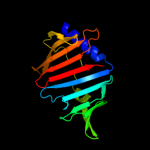
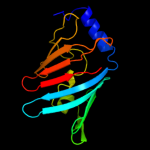
PDB 3bk5 chain A
Region: 24 - 189
Aligned: 166
Modelled: 166
Confidence: 99.2%
Identity: 12%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative outer membrane lipoprotein-sorting protein;
PDBTitle: crystal structure of putative outer membrane lipoprotein-sorting2 protein domain from vibrio parahaemolyticus
Phyre2
| 5 |
|
PDB 2v43 chain A
Region: 19 - 187
Aligned: 160
Modelled: 169
Confidence: 99.0%
Identity: 10%
PDB header:regulator
Chain: A: PDB Molecule:sigma-e factor regulatory protein rseb;
PDBTitle: crystal structure of rseb: a sensor for periplasmic stress2 response in e. coli
Phyre2
| 6 |
|
PDB 2yzy chain A
Region: 66 - 201
Aligned: 118
Modelled: 121
Confidence: 98.2%
Identity: 14%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative uncharacterized protein ttha1012;
PDBTitle: crystal structure of uncharacterized conserved protein from thermus2 thermophilus hb8
Phyre2
| 7 |
|
PDB 3mha chain B
Region: 18 - 177
Aligned: 158
Modelled: 159
Confidence: 58.3%
Identity: 13%
PDB header:lipid binding protein
Chain: B: PDB Molecule:lipoprotein lprg;
PDBTitle: crystal structure of lprg from mycobacterium tuberculosis bound to pim
Phyre2
| 8 |
|
PDB 2byo chain A domain 1
Region: 18 - 177
Aligned: 160
Modelled: 160
Confidence: 25.7%
Identity: 9%
Fold: LolA-like prokaryotic lipoproteins and lipoprotein localization factors
Superfamily: Prokaryotic lipoproteins and lipoprotein localization factors
Family: LppX-like
Phyre2
| 9 |
|
PDB 3gf5 chain A
Region: 189 - 203
Aligned: 15
Modelled: 15
Confidence: 13.1%
Identity: 27%
PDB header:structural protein
Chain: A: PDB Molecule:major vault protein;
PDBTitle: crystal structure of the p21 r1-r7 n-terminal domain of murine mvp
Phyre2
| 10 |
|
PDB 3r2c chain J
Region: 185 - 199
Aligned: 15
Modelled: 15
Confidence: 11.5%
Identity: 33%
PDB header:transcription/rna
Chain: J: PDB Molecule:30s ribosomal protein s10;
PDBTitle: crystal structure of antitermination factors nusb and nuse in complex2 with boxa rna
Phyre2
| 11 |
|
PDB 2f9j chain P
Region: 180 - 198
Aligned: 19
Modelled: 19
Confidence: 9.3%
Identity: 21%
PDB header:rna binding protein
Chain: P: PDB Molecule:splicing factor 3b subunit 1;
PDBTitle: 3.0 angstrom resolution structure of a y22m mutant of the spliceosomal2 protein p14 bound to a region of sf3b155
Phyre2
| 12 |
|
PDB 2jna chain A domain 1
Region: 1 - 22
Aligned: 22
Modelled: 22
Confidence: 9.1%
Identity: 41%
Fold: Dodecin subunit-like
Superfamily: YdgH-like
Family: YdgH-like
Phyre2
| 13 |
|
PDB 1o5u chain A
Region: 60 - 101
Aligned: 40
Modelled: 42
Confidence: 8.8%
Identity: 18%
Fold: Double-stranded beta-helix
Superfamily: RmlC-like cupins
Family: Hypothetical protein TM1112
Phyre2
| 14 |
|
PDB 1qub chain A domain 5
Region: 76 - 91
Aligned: 16
Modelled: 16
Confidence: 7.2%
Identity: 13%
Fold: Complement control module/SCR domain
Superfamily: Complement control module/SCR domain
Family: Complement control module/SCR domain
Phyre2
| 15 |
|
PDB 1c1z chain A domain 5
Region: 76 - 91
Aligned: 16
Modelled: 16
Confidence: 7.1%
Identity: 13%
Fold: Complement control module/SCR domain
Superfamily: Complement control module/SCR domain
Family: Complement control module/SCR domain
Phyre2
| 16 |
|
PDB 1yrr chain A domain 1
Region: 191 - 201
Aligned: 11
Modelled: 11
Confidence: 7.1%
Identity: 9%
Fold: Composite domain of metallo-dependent hydrolases
Superfamily: Composite domain of metallo-dependent hydrolases
Family: N-acetylglucosamine-6-phosphate deacetylase, NagA
Phyre2
| 17 |
|
PDB 1gwm chain A
Region: 132 - 197
Aligned: 64
Modelled: 66
Confidence: 6.1%
Identity: 11%
Fold: Galactose-binding domain-like
Superfamily: Galactose-binding domain-like
Family: Family 29 carbohydrate binding module, CBM29
Phyre2
| 18 |
|
PDB 1zak chain A domain 2
Region: 189 - 201
Aligned: 13
Modelled: 13
Confidence: 5.8%
Identity: 15%
Fold: Rubredoxin-like
Superfamily: Microbial and mitochondrial ADK, insert "zinc finger" domain
Family: Microbial and mitochondrial ADK, insert "zinc finger" domain
Phyre2
| 19 |
|
PDB 2ky8 chain A
Region: 180 - 194
Aligned: 15
Modelled: 15
Confidence: 5.7%
Identity: 33%
PDB header:transcription/dna
Chain: A: PDB Molecule:methyl-cpg-binding domain protein 2;
PDBTitle: solution structure and dynamic analysis of chicken mbd2 methyl binding2 domain bound to a target methylated dna sequence
Phyre2
| 20 |
|
PDB 1iwm chain A
Region: 19 - 177
Aligned: 158
Modelled: 159
Confidence: 5.6%
Identity: 10%
Fold: LolA-like prokaryotic lipoproteins and lipoprotein localization factors
Superfamily: Prokaryotic lipoproteins and lipoprotein localization factors
Family: Outer membrane lipoprotein receptor LolB
Phyre2