| 1 |
|
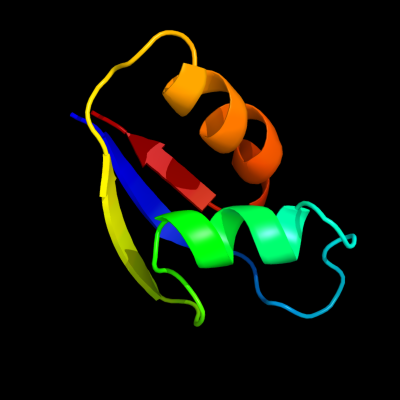
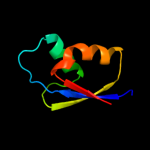
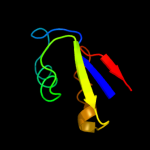
PDB 1bxy chain A
Region: 1 - 59
Aligned: 58
Modelled: 59
Confidence: 99.9%
Identity: 43%
Fold: Ribosomal protein L30p/L7e
Superfamily: Ribosomal protein L30p/L7e
Family: Ribosomal protein L30p/L7e
Phyre2
| 2 |
|
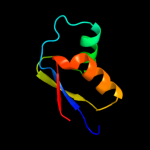
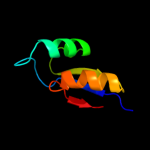
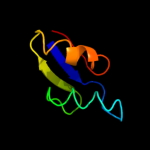
PDB 2gyc chain X domain 1
Region: 4 - 59
Aligned: 56
Modelled: 56
Confidence: 99.9%
Identity: 100%
Fold: Ribosomal protein L30p/L7e
Superfamily: Ribosomal protein L30p/L7e
Family: Ribosomal protein L30p/L7e
Phyre2
| 3 |
|
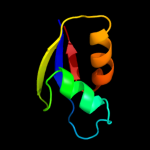
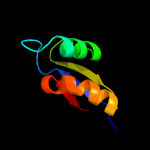
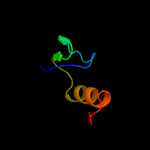
PDB 2zjr chain W domain 1
Region: 5 - 59
Aligned: 55
Modelled: 55
Confidence: 99.9%
Identity: 45%
Fold: Ribosomal protein L30p/L7e
Superfamily: Ribosomal protein L30p/L7e
Family: Ribosomal protein L30p/L7e
Phyre2
| 4 |
|
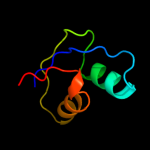
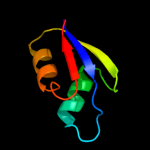
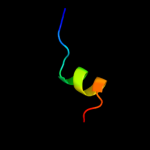
PDB 4a1c chain V
Region: 2 - 59
Aligned: 58
Modelled: 58
Confidence: 99.0%
Identity: 21%
PDB header:ribosome
Chain: V: PDB Molecule:60s ribosomal protein l7;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
Phyre2
| 5 |
|
PDB 3izc chain E
Region: 2 - 59
Aligned: 58
Modelled: 58
Confidence: 98.9%
Identity: 19%
PDB header:ribosome
Chain: E: PDB Molecule:60s ribosomal protein rpl11 (l5p);
PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
Phyre2
| 6 |
|
PDB 3iz5 chain E
Region: 2 - 59
Aligned: 58
Modelled: 58
Confidence: 98.9%
Identity: 28%
PDB header:ribosome
Chain: E: PDB Molecule:60s ribosomal protein l11 (l5p);
PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
Phyre2
| 7 |
|
PDB 1vqo chain W domain 1
Region: 4 - 59
Aligned: 56
Modelled: 56
Confidence: 98.9%
Identity: 25%
Fold: Ribosomal protein L30p/L7e
Superfamily: Ribosomal protein L30p/L7e
Family: Ribosomal protein L30p/L7e
Phyre2
| 8 |
|
PDB 3jyw chain F
Region: 2 - 59
Aligned: 58
Modelled: 58
Confidence: 98.9%
Identity: 19%
PDB header:ribosome
Chain: F: PDB Molecule:60s ribosomal protein l7(a);
PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
Phyre2
| 9 |
|
PDB 2zkr chain W
Region: 5 - 59
Aligned: 55
Modelled: 55
Confidence: 98.8%
Identity: 22%
PDB header:ribosomal protein/rna
Chain: W: PDB Molecule:rna expansion segment es10;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
Phyre2
| 10 |
|
PDB 1s1i chain F
Region: 5 - 59
Aligned: 55
Modelled: 55
Confidence: 98.7%
Identity: 18%
PDB header:ribosome
Chain: F: PDB Molecule:60s ribosomal protein l7-a;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
Phyre2
| 11 |
|
PDB 3exc chain X
Region: 19 - 59
Aligned: 41
Modelled: 41
Confidence: 11.6%
Identity: 24%
PDB header:hydrolase
Chain: X: PDB Molecule:uncharacterized protein;
PDBTitle: structure of the rna'se sso8090 from sulfolobus solfataricus
Phyre2
| 12 |
|
PDB 1jhf chain A domain 1
Region: 15 - 30
Aligned: 16
Modelled: 16
Confidence: 9.2%
Identity: 31%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: LexA repressor, N-terminal DNA-binding domain
Phyre2