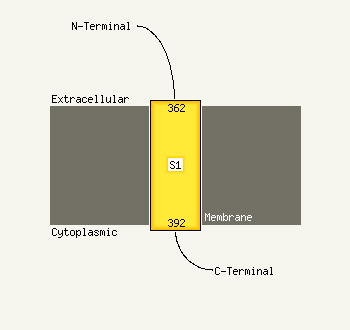
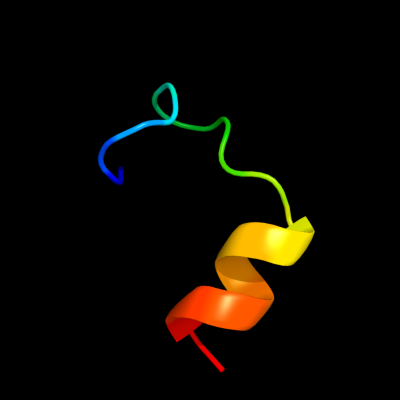
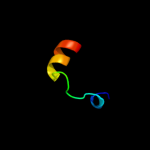
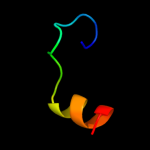
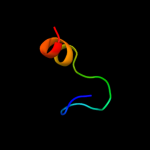
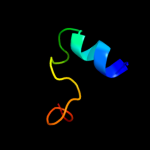
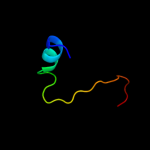
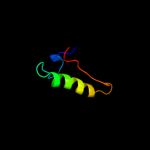
1 c2nscA_
40.8
25
PDB header: chaperoneChain: A: PDB Molecule: trigger factor;PDBTitle: structures of and interactions between domains of trigger factor from2 themotoga maritima
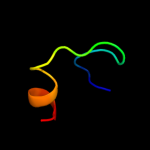
2 d1w26a2
35.3
20
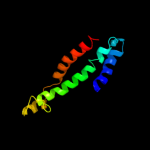
Fold: Ribosome binding domain-likeSuperfamily: Trigger factor ribosome-binding domainFamily: Trigger factor ribosome-binding domain3 d1p9ya_
34.3
20
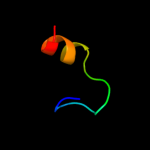
Fold: Ribosome binding domain-likeSuperfamily: Trigger factor ribosome-binding domainFamily: Trigger factor ribosome-binding domain4 d1t11a2
33.7
15
Fold: Ribosome binding domain-likeSuperfamily: Trigger factor ribosome-binding domainFamily: Trigger factor ribosome-binding domain5 c2d3o1_
30.3
25
PDB header: ribosomeChain: 1: PDB Molecule: trigger factor;PDBTitle: structure of ribosome binding domain of the trigger factor2 on the 50s ribosomal subunit from d. radiodurans
6 c2yvxD_
30.2
17
PDB header: transport proteinChain: D: PDB Molecule: mg2+ transporter mgte;PDBTitle: crystal structure of magnesium transporter mgte
7 c1t11A_
27.4
15
PDB header: chaperoneChain: A: PDB Molecule: trigger factor;PDBTitle: trigger factor
8 c2qgoA_
25.7
13
PDB header: biosynthetic proteinChain: A: PDB Molecule: putative fe-s biosynthesis protein;PDBTitle: crystal structure of a putative fe-s biosynthesis protein from2 lactobacillus acidophilus
9 d1osda_
21.7
21
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain10 d1cpza_
20.6
21
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain11 c2k2pA_
18.7
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atu1203;PDBTitle: solution nmr structure of protein atu1203 from agrobacterium2 tumefaciens. northeast structural genomics consortium (nesg) target3 att10, ontario center for structural proteomics target atc1183
12 c1yg0A_
18.7
24
PDB header: metal transportChain: A: PDB Molecule: cop associated protein;PDBTitle: solution structure of apo-copp from helicobacter pylori
13 d1mwza_
18.7
21
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain14 c1y3kA_
16.7
12
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the apo form of the fifth domain of2 menkes protein
15 c2ga7A_
16.3
18
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the copper(i) form of the third metal-2 binding domain of atp7a protein (menkes disease protein)
16 d1qb2a_
16.2
25
Fold: Signal peptide-binding domainSuperfamily: Signal peptide-binding domainFamily: Signal peptide-binding domain17 d2p2ea1
15.5
19
Fold: HesB-like domainSuperfamily: HesB-like domainFamily: HesB-like domain18 d1p6ta1
14.0
18
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain19 d1jw2a_
13.9
27
Fold: Open three-helical up-and-down bundleSuperfamily: Hemolysin expression modulating protein HHAFamily: Hemolysin expression modulating protein HHA20 c1vf5R_
13.5
26
PDB header: photosynthesisChain: R: PDB Molecule: protein pet l;PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
21 c2e75E_
not modelled
13.5
26
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex with 2-nonyl-4-2 hydroxyquinoline n-oxide (nqno) from m.laminosus
22 c2e74E_
not modelled
13.5
26
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex from m.laminosus
23 c2e76E_
not modelled
13.5
26
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
24 d2e74e1
not modelled
13.5
26
Fold: Single transmembrane helixSuperfamily: PetL subunit of the cytochrome b6f complexFamily: PetL subunit of the cytochrome b6f complex25 d1qzxa2
not modelled
13.2
25
Fold: Signal peptide-binding domainSuperfamily: Signal peptide-binding domainFamily: Signal peptide-binding domain26 c2ofhX_
not modelled
13.1
15
PDB header: hydrolase, membrane proteinChain: X: PDB Molecule: zinc-transporting atpase;PDBTitle: solution structure of the n-terminal domain of the zinc(ii) atpase2 ziaa in its apo form
27 c2aj1A_
not modelled
13.1
15
PDB header: hydrolaseChain: A: PDB Molecule: probable cadmium-transporting atpase;PDBTitle: solution structure of apocada
28 d2aw0a_
not modelled
12.6
26
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain29 d1w4ha1
not modelled
12.1
19
Fold: Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complexSuperfamily: Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complexFamily: Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex30 d1afia_
not modelled
11.7
18
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain31 c3bdlA_
not modelled
11.2
28
PDB header: hydrolaseChain: A: PDB Molecule: staphylococcal nuclease domain-containingPDBTitle: crystal structure of a truncated human tudor-sn
32 d1vyva2
not modelled
11.1
29
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases33 d2cyua1
not modelled
10.8
18
Fold: Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complexSuperfamily: Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complexFamily: Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex34 d2c4ka1
not modelled
10.7
27
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like35 c2kt2A_
not modelled
10.2
29
PDB header: oxidoreductaseChain: A: PDB Molecule: mercuric reductase;PDBTitle: structure of nmera, the n-terminal hma domain of tn501 mercuric2 reductase
36 d2ffha2
not modelled
9.9
29
Fold: Signal peptide-binding domainSuperfamily: Signal peptide-binding domainFamily: Signal peptide-binding domain37 c2kkhA_
not modelled
9.8
21
PDB header: metal transportChain: A: PDB Molecule: putative heavy metal transporter;PDBTitle: structure of the zinc binding domain of the atpase hma4
38 c1w26B_
not modelled
9.8
20
PDB header: chaperoneChain: B: PDB Molecule: trigger factor;PDBTitle: trigger factor in complex with the ribosome forms a2 molecular cradle for nascent proteins
39 c2iy3A_
not modelled
9.6
25
PDB header: rna-bindingChain: A: PDB Molecule: signal recognition particle protein ffh;PDBTitle: structure of the e. coli signal recognition particle2 bound to a translating ribosome
40 d1kvja_
not modelled
9.3
29
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain41 d1vs6z1
not modelled
9.3
25
Fold: L28p-likeSuperfamily: L28p-likeFamily: Ribosomal protein L31p42 c2vzkD_
not modelled
8.8
26
PDB header: transferaseChain: D: PDB Molecule: glutamate n-acetyltransferase 2 beta chain;PDBTitle: structure of the acyl-enzyme complex of an n-terminal2 nucleophile (ntn) hydrolase, oat2
43 c2oarA_
not modelled
8.6
18
PDB header: membrane proteinChain: A: PDB Molecule: large-conductance mechanosensitive channel;PDBTitle: mechanosensitive channel of large conductance (mscl)
44 d1sb6a_
not modelled
8.6
15
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain45 d1s6ua_
not modelled
8.4
35
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain46 d1uwda_
not modelled
8.4
25
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: PaaD-like47 d1p6ta2
not modelled
8.3
21
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain48 d2oara1
not modelled
7.5
18
Fold: Gated mechanosensitive channelSuperfamily: Gated mechanosensitive channelFamily: Gated mechanosensitive channel49 c3bvsA_
not modelled
7.4
10
PDB header: hydrolaseChain: A: PDB Molecule: alkylpurine dna glycosylase alkd;PDBTitle: crystal structure of bacillus cereus alkylpurine dna glycosylase alkd
50 c2kelB_
not modelled
7.2
29
PDB header: transcription repressorChain: B: PDB Molecule: uncharacterized protein 56b;PDBTitle: structure of the transcription regulator svtr from the2 hyperthermophilic archaeal virus sirv1
51 c1vf5E_
not modelled
7.2
23
PDB header: photosynthesisChain: E: PDB Molecule: protein pet l;PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
52 c1qzwC_
not modelled
7.1
25
PDB header: signaling protein/rnaChain: C: PDB Molecule: signal recognition 54 kda protein;PDBTitle: crystal structure of the complete core of archaeal srp and2 implications for inter-domain communication
53 c3k3gA_
not modelled
7.1
10
PDB header: transport proteinChain: A: PDB Molecule: urea transporter;PDBTitle: crystal structure of the urea transporter from desulfovibrio vulgaris2 bound to 1,3-dimethylurea
54 d2qifa1
not modelled
7.1
13
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain55 d3elga1
not modelled
7.0
19
Fold: BLIP-likeSuperfamily: BT0923-likeFamily: BT0923-like56 d1mn8a_
not modelled
7.0
21
Fold: Retroviral matrix proteinsSuperfamily: Retroviral matrix proteinsFamily: MMLV matrix protein-like57 d2aq0a1
not modelled
6.8
24
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Hef domain-like58 c2ksdA_
not modelled
6.6
10
PDB header: transferaseChain: A: PDB Molecule: aerobic respiration control sensor protein arcb;PDBTitle: backbone structure of the membrane domain of e. coli2 histidine kinase receptor arcb, center for structures of3 membrane proteins (csmp) target 4310c
59 d1ftra1
not modelled
6.1
24
Fold: Ferredoxin-likeSuperfamily: Formylmethanofuran:tetrahydromethanopterin formyltransferaseFamily: Formylmethanofuran:tetrahydromethanopterin formyltransferase60 d1t3la2
not modelled
5.6
29
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases61 d1nyta2
not modelled
5.3
33
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Shikimate dehydrogenase-like62 d3duea1
not modelled
5.1
13
Fold: BLIP-likeSuperfamily: BT0923-likeFamily: BT0923-like63 c2wwaJ_
not modelled
5.1
7
PDB header: ribosomeChain: J: PDB Molecule: 60s ribosomal protein l19;PDBTitle: cryo-em structure of idle yeast ssh1 complex bound to the2 yeast 80s ribosome
64 c3dv0I_
not modelled
5.1
13
PDB header: oxidoreductase/transferaseChain: I: PDB Molecule: dihydrolipoyllysine-residue acetyltransferasePDBTitle: snapshots of catalysis in the e1 subunit of the pyruvate2 dehydrogenase multi-enzyme complex