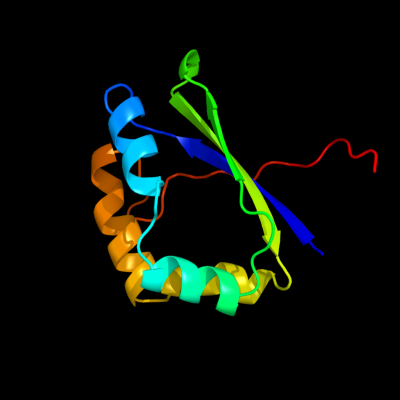
| 1 | d1x8da1
|
|
 |
100.0 |
100 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:YiiL-like |
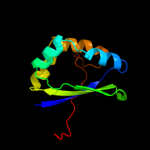
| 2 | c2qlwA_
|
|
 |
100.0 |
38 |
PDB header:isomerase
Chain: A: PDB Molecule:rhau;
PDBTitle: crystal structure of rhamnose mutarotase rhau of rhizobium2 leguminosarum
|
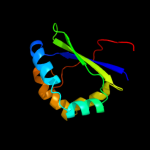
| 3 | c2qlxA_
|
|
 |
100.0 |
38 |
PDB header:isomerase
Chain: A: PDB Molecule:l-rhamnose mutarotase;
PDBTitle: crystal structure of rhamnose mutarotase rhau of rhizobium2 leguminosarum in complex with l-rhamnose
|
| 4 | c3bm7A_
|
|
 |
97.2 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein of unknown function with ferredoxin-like fold;
PDBTitle: crystal structure of a putative antibiotic biosynthesis monooxygenase2 (cc_2132) from caulobacter crescentus cb15 at 1.35 a resolution
|
| 5 | c2fb0A_
|
|
 |
97.1 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved hypothetical protein;
PDBTitle: crystal structure of conserved protein of unknown function from2 bacteroides thetaiotaomicron vpi-5482 at 2.10 a resolution, possible3 oxidoreductase
|
| 6 | c3e8oB_
|
|
 |
96.9 |
8 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:uncharacterized protein with erredoxin-like fold;
PDBTitle: crystal structure of a putative antibiotic biosynthesis monooxygenase2 (dr_2100) from deinococcus radiodurans at 1.40 a resolution
|
| 7 | d2omoa1
|
|
 |
96.8 |
15 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:PA3566-like |
| 8 | c3f44A_
|
|
 |
96.7 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative monooxygenase;
PDBTitle: crystal structure of putative monooxygenase (yp_193413.1) from2 lactobacillus acidophilus ncfm at 1.55 a resolution
|
| 9 | c2omoC_
|
|
 |
96.6 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:duf176;
PDBTitle: putative antibiotic biosynthesis monooxygenase from nitrosomonas2 europaea
|
| 10 | d1y0ha_
|
|
 |
96.6 |
10 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:PA3566-like |
| 11 | c2gffB_
|
|
 |
96.5 |
10 |
PDB header:sugar binding protein
Chain: B: PDB Molecule:lsrg protein;
PDBTitle: crystal structure of yersinia pestis lsrg
|
| 12 | c2bbeA_
|
|
 |
96.5 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein so0527;
PDBTitle: crystal structure of protein so0527 from shewanella oneidensis
|
| 13 | d2pd1a1
|
|
 |
96.3 |
10 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:PA3566-like |
| 14 | d1x7va_
|
|
 |
96.2 |
13 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:PA3566-like |
| 15 | c3gz7B_
|
|
 |
96.0 |
13 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:putative antibiotic biosynthesis monooxygenase;
PDBTitle: crystal structure of putative antibiotic biosynthesis2 monooxygenase (np_888398.1) from bordetella bronchiseptica3 at 2.15 a resolution
|
| 16 | c3mcsB_
|
|
 |
96.0 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative monooxygenase;
PDBTitle: crystal structure of putative monooxygenase (fn1347) from2 fusobacterium nucleatum subsp. nucleatum atcc 25586 at 2.55 a3 resolution
|
| 17 | c3fgvB_
|
|
 |
95.5 |
9 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:uncharacterized protein with ferredoxin-like fold;
PDBTitle: crystal structure of a putative antibiotic biosynthesis monooxygenase2 (spo2313) from silicibacter pomeroyi dss-3 at 1.30 a resolution
|
| 18 | c3kkfA_
|
|
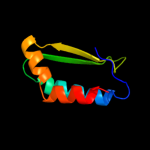 |
95.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative antibiotic biosynthesis monooxygenase;
PDBTitle: crystal structure of putative antibiotic biosynthesis2 monooxygenase (np_810307.1) from bacteriodes3 thetaiotaomicron vpi-5482 at 1.30 a resolution
|
| 19 | d1tuva_
|
|
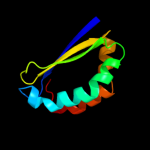 |
93.8 |
20 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:PA3566-like |
| 20 | d1iuja_
|
|
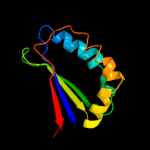 |
90.0 |
13 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:PG130-like |
| 21 | d1vqsa_ |
|
not modelled |
89.1 |
16 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:NIPSNAP |
| 22 | d1vqya1 |
|
not modelled |
87.3 |
11 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:NIPSNAP |
| 23 | c3kngA_ |
|
not modelled |
84.3 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:snoab;
PDBTitle: crystal structure of snoab, a cofactor-independent oxygenase2 from streptomyces nogalater, determined to 1.9 resolution
|
| 24 | d1q8ba_ |
|
not modelled |
70.9 |
18 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:Hypothetical protein YjcS |
| 25 | c3hx9B_ |
|
not modelled |
64.4 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:protein rv3592;
PDBTitle: structure of heme-degrader, mhud (rv3592), from2 mycobacterium tuberculosis with two hemes bound in its3 active site
|
| 26 | d2atza1 |
|
not modelled |
55.8 |
19 |
Fold:Prim-pol domain
Superfamily:Prim-pol domain
Family:HP0184-like |
| 27 | d1tuwa_ |
|
not modelled |
29.0 |
17 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:Polyketide synthesis cyclase |
| 28 | c3louB_ |
|
not modelled |
21.9 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of formyltetrahydrofolate deformylase (yp_105254.1)2 from burkholderia mallei atcc 23344 at 1.90 a resolution
|
| 29 | c3fj2A_ |
|
not modelled |
21.5 |
8 |
PDB header:unknown function
Chain: A: PDB Molecule:monooxygenase-like protein;
PDBTitle: crystal structure of a monooxygenase-like protein (lin2316) from2 listeria innocua at 1.85 a resolution
|
| 30 | d2cz4a1 |
|
not modelled |
20.5 |
33 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 31 | d2zdpa1 |
|
not modelled |
18.9 |
17 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:PG130-like |
| 32 | c2x7vA_ |
|
not modelled |
18.3 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable endonuclease 4;
PDBTitle: crystal structure of thermotoga maritima endonuclease iv in2 the presence of zinc
|
| 33 | c3thdD_ |
|
not modelled |
18.0 |
22 |
PDB header:hydrolase
Chain: D: PDB Molecule:beta-galactosidase;
PDBTitle: crystal structure of human beta-galactosidase in complex with 1-2 deoxygalactonojirimycin
|
| 34 | d2ns1b1 |
|
not modelled |
16.1 |
17 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 35 | c2jdjB_ |
|
not modelled |
12.2 |
5 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:redy-like protein;
PDBTitle: crystal structure of hapk from hahella chejuensis
|
| 36 | c2rodB_ |
|
not modelled |
11.7 |
33 |
PDB header:apoptosis
Chain: B: PDB Molecule:noxa;
PDBTitle: solution structure of mcl-1 complexed with noxaa
|
| 37 | c3obiC_ |
|
not modelled |
10.9 |
15 |
PDB header:hydrolase
Chain: C: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (np_949368)2 from rhodopseudomonas palustris cga009 at 1.95 a resolution
|
| 38 | c2qqdG_ |
|
not modelled |
10.8 |
31 |
PDB header:lyase
Chain: G: PDB Molecule:pyruvoyl-dependent arginine decarboxylase (ec
PDBTitle: n47a mutant of pyruvoyl-dependent arginine decarboxylase2 from methanococcus jannashii
|
| 39 | d1vfja_ |
|
not modelled |
10.3 |
11 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 40 | c3mhyC_ |
|
not modelled |
10.2 |
22 |
PDB header:signaling protein
Chain: C: PDB Molecule:pii-like protein pz;
PDBTitle: a new pii protein structure
|
| 41 | c3o8wA_ |
|
not modelled |
10.1 |
11 |
PDB header:signaling protein
Chain: A: PDB Molecule:nitrogen regulatory protein p-ii (glnb-1);
PDBTitle: archaeoglobus fulgidus glnk1
|
| 42 | d2hh8a1 |
|
not modelled |
10.1 |
21 |
Fold:YdfO-like
Superfamily:YdfO-like
Family:YdfO-like |
| 43 | c3hfkB_ |
|
not modelled |
9.8 |
22 |
PDB header:isomerase
Chain: B: PDB Molecule:4-methylmuconolactone methylisomerase;
PDBTitle: crystal structure of 4-methylmuconolactone methylisomerase2 (h52a) in complex with 4-methylmuconolactone
|
| 44 | c3d3aA_ |
|
not modelled |
9.7 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-galactosidase;
PDBTitle: crystal structure of a beta-galactosidase from bacteroides2 thetaiotaomicron
|
| 45 | c2hfqA_ |
|
not modelled |
9.4 |
33 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: nmr structure of protein ne1680 from nitrosomonas europaea:2 northeast structural genomics consortium target net5
|
| 46 | d2hfqa1 |
|
not modelled |
9.4 |
33 |
Fold:NE1680-like
Superfamily:NE1680-like
Family:NE1680-like |
| 47 | c2rd5D_ |
|
not modelled |
9.2 |
12 |
PDB header:protein binding
Chain: D: PDB Molecule:pii protein;
PDBTitle: structural basis for the regulation of n-acetylglutamate kinase by pii2 in arabidopsis thaliana
|
| 48 | c2pfuA_ |
|
not modelled |
9.1 |
13 |
PDB header:transport protein
Chain: A: PDB Molecule:biopolymer transport exbd protein;
PDBTitle: nmr strcuture determination of the periplasmic domain of exbd from2 e.coli
|
| 49 | d1tafa_ |
|
not modelled |
9.0 |
21 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:TBP-associated factors, TAFs |
| 50 | c3bzqA_ |
|
not modelled |
8.9 |
11 |
PDB header:signaling protein/transcription
Chain: A: PDB Molecule:nitrogen regulatory protein p-ii;
PDBTitle: high resolution crystal structure of nitrogen regulatory protein2 (rv2919c) of mycobacterium tuberculosis
|
| 51 | c3ncpD_ |
|
not modelled |
8.4 |
26 |
PDB header:signaling protein
Chain: D: PDB Molecule:nitrogen regulatory protein p-ii (glnb-2);
PDBTitle: glnk2 from archaeoglobus fulgidus
|
| 52 | d1qy7a_ |
|
not modelled |
8.1 |
17 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 53 | c2lf3A_ |
|
not modelled |
7.6 |
28 |
PDB header:signaling protein
Chain: A: PDB Molecule:effector protein hopab3;
PDBTitle: solution nmr structure of hoppmal_281_385 from pseudomonas syringae2 pv. maculicola str. es4326, midwest center for structural genomics3 target apc40104.5 and northeast structural genomics consortium target4 pst2a
|
| 54 | c3ogrA_ |
|
not modelled |
7.5 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-galactosidase;
PDBTitle: complex structure of beta-galactosidase from trichoderma reesei with2 galactose
|
| 55 | c1xc6A_ |
|
not modelled |
7.2 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-galactosidase;
PDBTitle: native structure of beta-galactosidase from penicillium sp. in complex2 with galactose
|
| 56 | c2k42A_ |
|
not modelled |
7.1 |
19 |
PDB header:signaling protein
Chain: A: PDB Molecule:wiskott-aldrich syndrome protein;
PDBTitle: solution structure of the gtpase binding domain of wasp in2 complex with espfu, an ehec effector
|
| 57 | d2piia_ |
|
not modelled |
7.0 |
11 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 58 | c1ceeB_ |
|
not modelled |
7.0 |
18 |
PDB header:structural protein regulation
Chain: B: PDB Molecule:wiskott-aldrich syndrome protein wasp;
PDBTitle: solution structure of cdc42 in complex with the gtpase2 binding domain of wasp
|
| 59 | c2dclB_ |
|
not modelled |
6.6 |
17 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical upf0166 protein ph1503;
PDBTitle: structure of ph1503 protein from pyrococcus horikoshii ot3
|
| 60 | c3s8sA_ |
|
not modelled |
6.6 |
21 |
PDB header:transcription
Chain: A: PDB Molecule:histone-lysine n-methyltransferase setd1a;
PDBTitle: crystal structure of the rrm domain of human setd1a
|
| 61 | c2j9dG_ |
|
not modelled |
6.0 |
11 |
PDB header:membrane transport
Chain: G: PDB Molecule:hypothetical nitrogen regulatory pii-like
PDBTitle: structure of glnk1 with bound effectors indicates2 regulatory mechanism for ammonia uptake
|
| 62 | d1gx5a_ |
|
not modelled |
5.7 |
11 |
Fold:DNA/RNA polymerases
Superfamily:DNA/RNA polymerases
Family:RNA-dependent RNA-polymerase |
| 63 | c3n0vD_ |
|
not modelled |
5.7 |
15 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
|
| 64 | d1hwua_ |
|
not modelled |
5.5 |
16 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 65 | c3mjkE_ |
|
not modelled |
5.2 |
20 |
PDB header:hormone
Chain: E: PDB Molecule:platelet-derived growth factor subunit a;
PDBTitle: structure of a growth factor precursor
|
| 66 | c3nrbD_ |
|
not modelled |
5.2 |
15 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (puru,2 pp_1943) from pseudomonas putida kt2440 at 2.05 a resolution
|
| 67 | d1p42a1 |
|
not modelled |
5.1 |
20 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase LpxC |
| 68 | d1qw1a1 |
|
not modelled |
5.1 |
8 |
Fold:SH3-like barrel
Superfamily:C-terminal domain of transcriptional repressors
Family:FeoA-like |