| 1 |
|
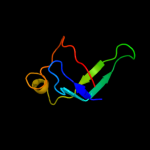
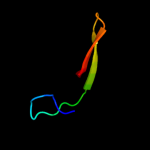
PDB 3gt2 chain A
Region: 32 - 54
Aligned: 23
Modelled: 23
Confidence: 25.0%
Identity: 35%
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the p60 domain from m. avium2 paratuberculosis antigen map1272c
Phyre2
| 2 |
|
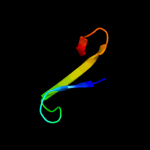
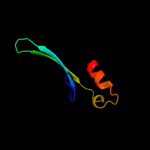
PDB 2yxy chain A
Region: 130 - 155
Aligned: 21
Modelled: 26
Confidence: 16.8%
Identity: 48%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical conserved protein, gk0453;
PDBTitle: crystarl structure of hypothetical conserved protein, gk0453
Phyre2
| 3 |
|
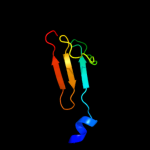
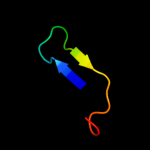
PDB 3ipj chain B
Region: 107 - 126
Aligned: 20
Modelled: 20
Confidence: 16.4%
Identity: 30%
PDB header:transferase
Chain: B: PDB Molecule:pts system, iiabc component;
PDBTitle: the crystal structure of one domain of the pts system, iiabc component2 from clostridium difficile
Phyre2
| 4 |
|
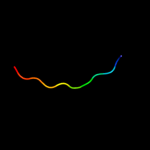
PDB 2eqn chain A
Region: 72 - 122
Aligned: 50
Modelled: 51
Confidence: 14.8%
Identity: 16%
PDB header:transcription
Chain: A: PDB Molecule:hypothetical protein loc92345;
PDBTitle: solution structure of the naf1 domain of hypothetical2 protein bc008207 [homo sapiens]
Phyre2
| 5 |
|
PDB 3pbi chain A
Region: 32 - 54
Aligned: 23
Modelled: 23
Confidence: 14.7%
Identity: 30%
PDB header:hydrolase
Chain: A: PDB Molecule:invasion protein;
PDBTitle: structure of the peptidoglycan hydrolase ripb (rv1478) from2 mycobacterium tuberculosis at 1.6 resolution
Phyre2
| 6 |
|
PDB 1iba chain A
Region: 107 - 126
Aligned: 20
Modelled: 20
Confidence: 8.7%
Identity: 35%
PDB header:phoshphotransferase
Chain: A: PDB Molecule:glucose permease;
PDBTitle: glucose permease (domain iib), nmr, 11 structures
Phyre2
| 7 |
|
PDB 3e8v chain A
Region: 124 - 141
Aligned: 18
Modelled: 18
Confidence: 8.6%
Identity: 28%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:possible transglutaminase-family protein;
PDBTitle: crystal structure of a possible transglutaminase-family2 protein proteolytic fragment from bacteroides fragilis
Phyre2
| 8 |
|
PDB 2xiv chain A
Region: 25 - 54
Aligned: 30
Modelled: 28
Confidence: 8.4%
Identity: 27%
PDB header:structural protein
Chain: A: PDB Molecule:hypothetical invasion protein;
PDBTitle: structure of rv1477, hypothetical invasion protein of2 mycobacterium tuberculosis
Phyre2
| 9 |
|
PDB 2yuj chain A
Region: 79 - 86
Aligned: 8
Modelled: 8
Confidence: 7.7%
Identity: 63%
PDB header:protein binding
Chain: A: PDB Molecule:ubiquitin fusion degradation 1-like;
PDBTitle: solution structure of human ubiquitin fusion degradation2 protein 1 homolog ufd1
Phyre2
| 10 |
|
PDB 1drs chain A
Region: 34 - 70
Aligned: 37
Modelled: 37
Confidence: 7.7%
Identity: 32%
Fold: Snake toxin-like
Superfamily: Snake toxin-like
Family: Dendroaspin
Phyre2
| 11 |
|
PDB 3npf chain B
Region: 25 - 54
Aligned: 30
Modelled: 30
Confidence: 7.0%
Identity: 47%
PDB header:hydrolase
Chain: B: PDB Molecule:putative dipeptidyl-peptidase vi;
PDBTitle: crystal structure of a putative dipeptidyl-peptidase vi (bacova_00612)2 from bacteroides ovatus at 1.72 a resolution
Phyre2
| 12 |
|
PDB 1d0n chain A domain 6
Region: 106 - 160
Aligned: 55
Modelled: 55
Confidence: 7.0%
Identity: 24%
Fold: Gelsolin-like
Superfamily: Actin depolymerizing proteins
Family: Gelsolin-like
Phyre2
| 13 |
|
PDB 1zc1 chain A
Region: 76 - 86
Aligned: 11
Modelled: 11
Confidence: 6.5%
Identity: 36%
PDB header:protein turnover
Chain: A: PDB Molecule:ubiquitin fusion degradation protein 1;
PDBTitle: ufd1 exhibits the aaa-atpase fold with two distinct2 ubiquitin interaction sites
Phyre2
| 14 |
|
PDB 1sf9 chain A
Region: 130 - 155
Aligned: 21
Modelled: 26
Confidence: 6.4%
Identity: 43%
Fold: SH3-like barrel
Superfamily: Hypothetical protein YfhH
Family: Hypothetical protein YfhH
Phyre2
| 15 |
|
PDB 1ik9 chain C
Region: 135 - 156
Aligned: 21
Modelled: 22
Confidence: 5.8%
Identity: 57%
PDB header:gene regulation/ligase
Chain: C: PDB Molecule:dna ligase iv;
PDBTitle: crystal structure of a xrcc4-dna ligase iv complex
Phyre2
| 16 |
|
PDB 1gjw chain A domain 1
Region: 155 - 160
Aligned: 6
Modelled: 6
Confidence: 5.7%
Identity: 83%
Fold: Glycosyl hydrolase domain
Superfamily: Glycosyl hydrolase domain
Family: alpha-Amylases, C-terminal beta-sheet domain
Phyre2
| 17 |
|
PDB 1ig8 chain A domain 1
Region: 110 - 159
Aligned: 50
Modelled: 50
Confidence: 5.5%
Identity: 10%
Fold: Ribonuclease H-like motif
Superfamily: Actin-like ATPase domain
Family: Hexokinase
Phyre2
| 18 |
|
PDB 2esl chain A domain 1
Region: 80 - 95
Aligned: 16
Modelled: 16
Confidence: 5.1%
Identity: 25%
Fold: Cyclophilin-like
Superfamily: Cyclophilin-like
Family: Cyclophilin (peptidylprolyl isomerase)
Phyre2
| 19 |
|
PDB 1tif chain A
Region: 119 - 138
Aligned: 20
Modelled: 20
Confidence: 5.1%
Identity: 25%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: Translation initiation factor IF3, N-terminal domain
Family: Translation initiation factor IF3, N-terminal domain
Phyre2
| 20 |
|
PDB 2kmf chain A
Region: 148 - 159
Aligned: 12
Modelled: 12
Confidence: 5.0%
Identity: 17%
PDB header:photosynthesis
Chain: A: PDB Molecule:photosystem ii 11 kda protein;
PDBTitle: solution structure of psb27 from cyanobacterial photosystem2 ii
Phyre2