| 1 | d3b60a2
|
|
 |
99.9 |
11 |
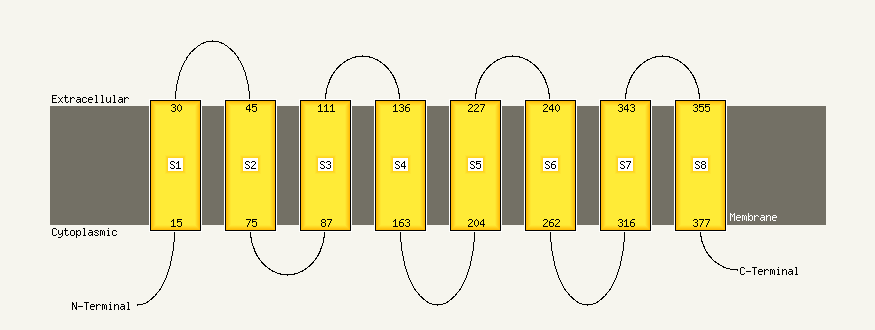
Fold:ABC transporter transmembrane region
Superfamily:ABC transporter transmembrane region
Family:ABC transporter transmembrane region |
| 2 | c2yl4A_
|
|
 |
99.9 |
11 |
PDB header:membrane protein
Chain: A: PDB Molecule:atp-binding cassette sub-family b member 10,
PDBTitle: structure of the human mitochondrial abc transporter, abcb10
|
| 3 | d2hyda2
|
|
 |
99.9 |
10 |
Fold:ABC transporter transmembrane region
Superfamily:ABC transporter transmembrane region
Family:ABC transporter transmembrane region |
| 4 | c2hydB_
|
|
 |
99.9 |
10 |
PDB header:transport protein
Chain: B: PDB Molecule:abc transporter homolog;
PDBTitle: multidrug abc transporter sav1866
|
| 5 | c3g5uB_
|
|
 |
99.9 |
8 |
PDB header:membrane protein
Chain: B: PDB Molecule:multidrug resistance protein 1a;
PDBTitle: structure of p-glycoprotein reveals a molecular basis for2 poly-specific drug binding
|
| 6 | c3b5wE_
|
|
 |
99.8 |
13 |
PDB header:membrane protein
Chain: E: PDB Molecule:lipid a export atp-binding/permease protein msba;
PDBTitle: crystal structure of eschericia coli msba
|
| 7 | d1pf4a2
|
|
 |
99.6 |
13 |
Fold:ABC transporter transmembrane region
Superfamily:ABC transporter transmembrane region
Family:ABC transporter transmembrane region |
| 8 | c3b5xB_
|
|
 |
99.6 |
11 |
PDB header:membrane protein
Chain: B: PDB Molecule:lipid a export atp-binding/permease protein msba;
PDBTitle: crystal structure of msba from vibrio cholerae
|
| 9 | d1ou5a1
|
|
 |
63.8 |
42 |
Fold:Poly A polymerase C-terminal region-like
Superfamily:Poly A polymerase C-terminal region-like
Family:Poly A polymerase C-terminal region-like |
| 10 | d1miwa1
|
|
 |
21.2 |
33 |
Fold:Poly A polymerase C-terminal region-like
Superfamily:Poly A polymerase C-terminal region-like
Family:Poly A polymerase C-terminal region-like |
| 11 | d1n2aa1
|
|
 |
19.4 |
9 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 12 | c3gehA_
|
|
 |
18.3 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:trna modification gtpase mnme;
PDBTitle: crystal structure of mnme from nostoc in complex with gdp, folinic2 acid and zn
|
| 13 | d1aw9a1
|
|
 |
18.2 |
11 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 14 | d2gsqa1
|
|
 |
17.1 |
11 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 15 | c1m0uB_
|
|
 |
17.1 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:gst2 gene product;
PDBTitle: crystal structure of the drosophila glutathione s-2 transferase-2 in complex with glutathione
|
| 16 | c3a0hk_
|
|
 |
16.0 |
50 |
PDB header:electron transport
Chain: K: PDB Molecule:photosystem ii reaction center protein k;
PDBTitle: crystal structure of i-substituted photosystem ii complex
|
| 17 | d2gsta1
|
|
 |
15.5 |
14 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 18 | d1hiob_
|
|
 |
14.3 |
11 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 19 | c1vheA_
|
|
 |
13.7 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:aminopeptidase/glucanase homolog;
PDBTitle: crystal structure of a aminopeptidase/glucanase homolog
|
| 20 | c2uz8A_
|
|
 |
12.8 |
7 |
PDB header:rna-binding protein
Chain: A: PDB Molecule:eukaryotic translation elongation factor 1
PDBTitle: the crystal structure of p18, human translation elongation2 factor 1 epsilon 1
|
| 21 | d1p3mh_ |
|
not modelled |
12.8 |
11 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 22 | c1aw9A_ |
|
not modelled |
12.2 |
7 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase iii;
PDBTitle: structure of glutathione s-transferase iii in apo form
|
| 23 | d1tw9a1 |
|
not modelled |
12.2 |
9 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 24 | d1iwpa_ |
|
not modelled |
11.7 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:Cobalamin (vitamin B12)-dependent enzymes
Family:Diol dehydratase, alpha subunit |
| 25 | c2hsmA_ |
|
not modelled |
10.8 |
8 |
PDB header:ligase/rna binding protein
Chain: A: PDB Molecule:glutamyl-trna synthetase, cytoplasmic;
PDBTitle: structural basis of yeast aminoacyl-trna synthetase complex2 formation revealed by crystal structures of two binary sub-3 complexes
|
| 26 | c1z23A_ |
|
not modelled |
10.5 |
26 |
PDB header:cell adhesion
Chain: A: PDB Molecule:crk-associated substrate;
PDBTitle: the serine-rich domain from crk-associated substrate2 (p130cas)
|
| 27 | c2l81A_ |
|
not modelled |
10.1 |
21 |
PDB header:cell adhesion
Chain: A: PDB Molecule:enhancer of filamentation 1;
PDBTitle: solution nmr structure of the serine-rich domain of hef1 (enhancer of2 filamentation 1) from homo sapiens, northeast structural genomics3 consortium target hr5554a
|
| 28 | c3a0bK_ |
|
not modelled |
10.1 |
50 |
PDB header:electron transport
Chain: K: PDB Molecule:photosystem ii reaction center protein k;
PDBTitle: crystal structure of br-substituted photosystem ii complex
|
| 29 | c3a0bk_ |
|
not modelled |
10.1 |
50 |
PDB header:electron transport
Chain: K: PDB Molecule:photosystem ii reaction center protein k;
PDBTitle: crystal structure of br-substituted photosystem ii complex
|
| 30 | d1eexa_ |
|
not modelled |
9.7 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Cobalamin (vitamin B12)-dependent enzymes
Family:Diol dehydratase, alpha subunit |
| 31 | d1b4pa1 |
|
not modelled |
9.7 |
12 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 32 | d1wspa1 |
|
not modelled |
8.6 |
36 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:DIX domain |
| 33 | d1id3d_ |
|
not modelled |
8.5 |
11 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 34 | c2qx2A_ |
|
not modelled |
7.4 |
3 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:sex pheromone staph-cam373;
PDBTitle: structure of the c-terminal domain of sex pheromone staph-cam3732 precursor from staphylococcus aureus
|
| 35 | c1byeA_ |
|
not modelled |
7.0 |
7 |
PDB header:transferase
Chain: A: PDB Molecule:protein (glutathione s-transferase);
PDBTitle: glutathione s-transferase i from mais in complex with2 atrazine glutathione conjugate
|
| 36 | c2yruA_ |
|
not modelled |
6.6 |
19 |
PDB header:apoptosis
Chain: A: PDB Molecule:steroid receptor rna activator 1;
PDBTitle: solution structure of mouse steroid receptor rna activator2 1 (sra1) protein
|
| 37 | d1pd3a_ |
|
not modelled |
6.4 |
18 |
Fold:ROP-like
Superfamily:Nonstructural protein ns2, Nep, M1-binding domain
Family:Nonstructural protein ns2, Nep, M1-binding domain |
| 38 | d1tzyb_ |
|
not modelled |
6.1 |
11 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 39 | c3lszA_ |
|
not modelled |
5.7 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase;
PDBTitle: crystal structure of glutathione s-transferase from2 rhodobacter sphaeroides
|
| 40 | d1b8xa1 |
|
not modelled |
5.4 |
13 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 41 | d1tu7a1 |
|
not modelled |
5.4 |
9 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 42 | c2akfB_ |
|
not modelled |
5.3 |
18 |
PDB header:protein binding
Chain: B: PDB Molecule:coronin-1a;
PDBTitle: crystal structure of the coiled-coil domain of coronin 1
|
| 43 | c2akfA_ |
|
not modelled |
5.3 |
18 |
PDB header:protein binding
Chain: A: PDB Molecule:coronin-1a;
PDBTitle: crystal structure of the coiled-coil domain of coronin 1
|
| 44 | c2akfC_ |
|
not modelled |
5.3 |
18 |
PDB header:protein binding
Chain: C: PDB Molecule:coronin-1a;
PDBTitle: crystal structure of the coiled-coil domain of coronin 1
|
| 45 | d2axtk1 |
|
not modelled |
5.3 |
50 |
Fold:Single transmembrane helix
Superfamily:Photosystem II reaction center protein K, PsbK
Family:PsbK-like |
| 46 | c2fq8A_ |
|
not modelled |
5.2 |
36 |
PDB header:unknown function
Chain: A: PDB Molecule:2f;
PDBTitle: nmr structure of 2f associated with lipid disc
|
| 47 | c2fq5A_ |
|
not modelled |
5.2 |
36 |
PDB header:unknown function
Chain: A: PDB Molecule:peptide 2f;
PDBTitle: nmr structure of 2f associated with lipid disc
|
| 48 | c1zvaA_ |
|
not modelled |
5.2 |
14 |
PDB header:viral protein
Chain: A: PDB Molecule:e2 glycoprotein;
PDBTitle: a structure-based mechanism of sars virus membrane fusion
|
| 49 | c1ou5A_ |
|
not modelled |
5.1 |
54 |
PDB header:translation, transferase
Chain: A: PDB Molecule:trna cca-adding enzyme;
PDBTitle: crystal structure of human cca-adding enzyme
|
| 50 | d1gnwa1 |
|
not modelled |
5.0 |
16 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |