| 1 |
|
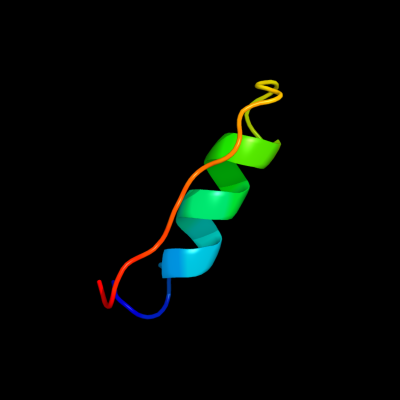
PDB 1wih chain A
Region: 58 - 83
Aligned: 26
Modelled: 26
Confidence: 10.4%
Identity: 27%
Fold: RRF/tRNA synthetase additional domain-like
Superfamily: Ribosome recycling factor, RRF
Family: Ribosome recycling factor, RRF
Phyre2
| 2 |
|
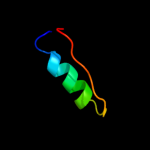
PDB 1xco chain A
Region: 36 - 69
Aligned: 31
Modelled: 34
Confidence: 9.9%
Identity: 29%
Fold: Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily: Isocitrate/Isopropylmalate dehydrogenase-like
Family: Phosphotransacetylase
Phyre2
| 3 |
|
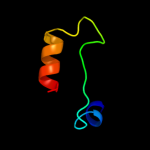
PDB 1r5j chain A
Region: 36 - 69
Aligned: 31
Modelled: 34
Confidence: 9.8%
Identity: 29%
Fold: Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily: Isocitrate/Isopropylmalate dehydrogenase-like
Family: Phosphotransacetylase
Phyre2
| 4 |
|
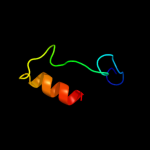
PDB 3ke2 chain A
Region: 63 - 83
Aligned: 21
Modelled: 21
Confidence: 9.3%
Identity: 52%
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein yp_928783.1;
PDBTitle: crystal structure of a duf2131 family protein (sama_2911) from2 shewanella amazonensis sb2b at 2.50 a resolution
Phyre2
| 5 |
|
PDB 2k9i chain B
Region: 12 - 30
Aligned: 17
Modelled: 19
Confidence: 8.9%
Identity: 65%
PDB header:dna binding protein
Chain: B: PDB Molecule:plasmid prn1, complete sequence;
PDBTitle: nmr structure of plasmid copy control protein orf56 from2 sulfolobus islandicus
Phyre2
| 6 |
|
PDB 3ogi chain C
Region: 9 - 32
Aligned: 24
Modelled: 24
Confidence: 8.0%
Identity: 50%
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:putative esat-6-like protein 6;
PDBTitle: crystal structure of the mycobacterium tuberculosis h37rv esxop2 complex (rv2346c-rv2347c)
Phyre2
| 7 |
|
PDB 1wqg chain A domain 1
Region: 58 - 83
Aligned: 26
Modelled: 26
Confidence: 7.9%
Identity: 27%
Fold: RRF/tRNA synthetase additional domain-like
Superfamily: Ribosome recycling factor, RRF
Family: Ribosome recycling factor, RRF
Phyre2
| 8 |
|
PDB 1jqn chain A
Region: 32 - 70
Aligned: 39
Modelled: 39
Confidence: 7.7%
Identity: 13%
Fold: TIM beta/alpha-barrel
Superfamily: Phosphoenolpyruvate/pyruvate domain
Family: Phosphoenolpyruvate carboxylase
Phyre2
| 9 |
|
PDB 2hzs chain L
Region: 74 - 83
Aligned: 10
Modelled: 10
Confidence: 7.1%
Identity: 70%
PDB header:transcription
Chain: L: PDB Molecule:rna polymerase ii mediator complex subunit 8;
PDBTitle: structure of the mediator head submodule med8c/18/20
Phyre2
| 10 |
|
PDB 2hzs chain J
Region: 74 - 83
Aligned: 10
Modelled: 10
Confidence: 7.1%
Identity: 70%
PDB header:transcription
Chain: J: PDB Molecule:rna polymerase ii mediator complex subunit 8;
PDBTitle: structure of the mediator head submodule med8c/18/20
Phyre2
| 11 |
|
PDB 1yl7 chain A domain 1
Region: 48 - 72
Aligned: 24
Modelled: 25
Confidence: 6.9%
Identity: 33%
Fold: NAD(P)-binding Rossmann-fold domains
Superfamily: NAD(P)-binding Rossmann-fold domains
Family: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
Phyre2
| 12 |
|
PDB 1jqo chain A
Region: 32 - 70
Aligned: 39
Modelled: 39
Confidence: 6.9%
Identity: 26%
PDB header:lyase
Chain: A: PDB Molecule:phosphoenolpyruvate carboxylase;
PDBTitle: crystal structure of c4-form phosphoenolpyruvate carboxylase from2 maize
Phyre2
| 13 |
|
PDB 1jqo chain A
Region: 32 - 70
Aligned: 39
Modelled: 39
Confidence: 6.9%
Identity: 26%
Fold: TIM beta/alpha-barrel
Superfamily: Phosphoenolpyruvate/pyruvate domain
Family: Phosphoenolpyruvate carboxylase
Phyre2