| 1 |
|
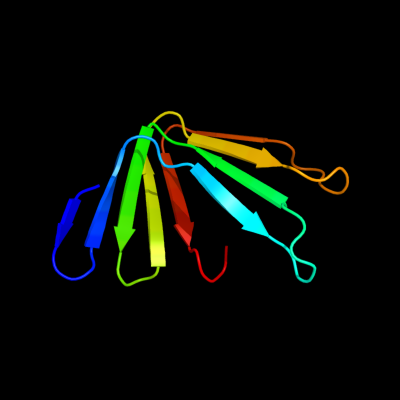
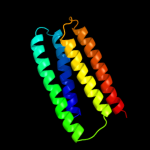
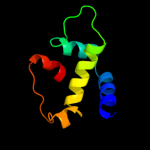
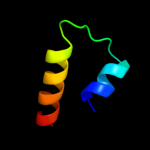
PDB 3my2 chain A
Region: 163 - 235
Aligned: 73
Modelled: 73
Confidence: 50.9%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:lipopolysaccharide export system protein lptc;
PDBTitle: crystal structure of lptc
Phyre2
| 2 |
|
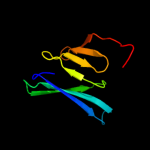
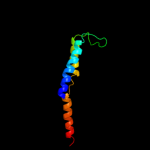
PDB 2rdc chain A
Region: 4 - 15
Aligned: 12
Modelled: 12
Confidence: 14.4%
Identity: 42%
PDB header:lipid binding protein
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative lipid binding protein (gsu0061) from2 geobacter sulfurreducens pca at 1.80 a resolution
Phyre2
| 3 |
|
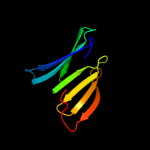
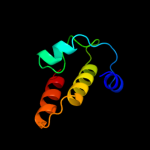
PDB 1utc chain A domain 2
Region: 156 - 234
Aligned: 77
Modelled: 79
Confidence: 12.6%
Identity: 17%
Fold: 7-bladed beta-propeller
Superfamily: Clathrin heavy-chain terminal domain
Family: Clathrin heavy-chain terminal domain
Phyre2
| 4 |
|
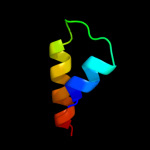
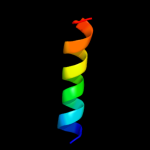
PDB 1oed chain A
Region: 10 - 127
Aligned: 118
Modelled: 118
Confidence: 10.3%
Identity: 7%
Fold: Neurotransmitter-gated ion-channel transmembrane pore
Superfamily: Neurotransmitter-gated ion-channel transmembrane pore
Family: Neurotransmitter-gated ion-channel transmembrane pore
Phyre2
| 5 |
|
PDB 2voy chain H
Region: 48 - 90
Aligned: 43
Modelled: 43
Confidence: 8.8%
Identity: 14%
PDB header:hydrolase
Chain: H: PDB Molecule:sarcoplasmic/endoplasmic reticulum calcium
PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
Phyre2
| 6 |
|
PDB 1c9l chain A
Region: 156 - 228
Aligned: 71
Modelled: 73
Confidence: 8.6%
Identity: 15%
PDB header:endocytosis/exocytosis
Chain: A: PDB Molecule:clathrin;
PDBTitle: peptide-in-groove interactions link target proteins to the2 b-propeller of clathrin
Phyre2
| 7 |
|
PDB 1bpo chain A
Region: 156 - 234
Aligned: 77
Modelled: 79
Confidence: 8.5%
Identity: 17%
PDB header:membrane protein
Chain: A: PDB Molecule:protein (clathrin);
PDBTitle: clathrin heavy-chain terminal domain and linker
Phyre2
| 8 |
|
PDB 1f5q chain B domain 1
Region: 5 - 35
Aligned: 31
Modelled: 31
Confidence: 8.0%
Identity: 19%
Fold: Cyclin-like
Superfamily: Cyclin-like
Family: Cyclin
Phyre2
| 9 |
|
PDB 1j1u chain A
Region: 65 - 136
Aligned: 67
Modelled: 72
Confidence: 7.0%
Identity: 18%
Fold: Adenine nucleotide alpha hydrolase-like
Superfamily: Nucleotidylyl transferase
Family: Class I aminoacyl-tRNA synthetases (RS), catalytic domain
Phyre2
| 10 |
|
PDB 1mhs chain A
Region: 11 - 87
Aligned: 69
Modelled: 77
Confidence: 7.0%
Identity: 16%
PDB header:membrane protein, proton transport
Chain: A: PDB Molecule:plasma membrane atpase;
PDBTitle: model of neurospora crassa proton atpase
Phyre2
| 11 |
|
PDB 1cii chain A
Region: 12 - 80
Aligned: 68
Modelled: 69
Confidence: 6.8%
Identity: 9%
PDB header:transmembrane protein
Chain: A: PDB Molecule:colicin ia;
PDBTitle: colicin ia
Phyre2
| 12 |
|
PDB 1rut chain X domain 1
Region: 5 - 13
Aligned: 9
Modelled: 9
Confidence: 5.8%
Identity: 22%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: LIM domain
Phyre2
| 13 |
|
PDB 3h3h chain A
Region: 128 - 195
Aligned: 66
Modelled: 68
Confidence: 5.8%
Identity: 14%
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized snoal-like protein;
PDBTitle: crystal structure of a snoal-like protein of unknown function2 (bth_ii0226) from burkholderia thailandensis e264 at 1.60 a3 resolution
Phyre2
| 14 |
|
PDB 2rdd chain B
Region: 339 - 358
Aligned: 20
Modelled: 20
Confidence: 5.6%
Identity: 25%
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:upf0092 membrane protein yajc;
PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
Phyre2
| 15 |
|
PDB 1g3n chain C domain 1
Region: 5 - 35
Aligned: 31
Modelled: 31
Confidence: 5.4%
Identity: 16%
Fold: Cyclin-like
Superfamily: Cyclin-like
Family: Cyclin
Phyre2
| 16 |
|
PDB 2cch chain B domain 1
Region: 5 - 35
Aligned: 31
Modelled: 31
Confidence: 5.3%
Identity: 16%
Fold: Cyclin-like
Superfamily: Cyclin-like
Family: Cyclin
Phyre2
| 17 |
|
PDB 1b0n chain A domain 1
Region: 93 - 102
Aligned: 10
Modelled: 10
Confidence: 5.3%
Identity: 30%
Fold: Dimerisation interlock
Superfamily: SinR repressor dimerisation domain-like
Family: SinR repressor dimerisation domain-like
Phyre2
| 18 |
|
PDB 3jvo chain A
Region: 69 - 88
Aligned: 20
Modelled: 20
Confidence: 5.2%
Identity: 20%
PDB header:viral protein
Chain: A: PDB Molecule:gp6;
PDBTitle: crystal structure of bacteriophage hk97 gp6
Phyre2