| 1 |
|
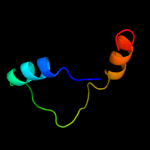
PDB 2gu0 chain A
Region: 8 - 94
Aligned: 67
Modelled: 67
Confidence: 41.6%
Identity: 24%
PDB header:viral protein
Chain: A: PDB Molecule:nonstructural protein 2;
PDBTitle: crystal structure of human rotavirus nsp2 (group c /2 bristol strain)
Phyre2
| 2 |
|
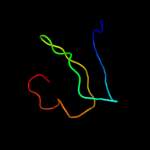
PDB 1s5q chain B
Region: 2 - 66
Aligned: 62
Modelled: 65
Confidence: 29.9%
Identity: 21%
Fold: PAH2 domain
Superfamily: PAH2 domain
Family: PAH2 domain
Phyre2
| 3 |
|
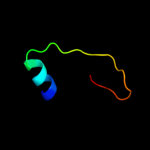
PDB 2f05 chain A domain 1
Region: 2 - 66
Aligned: 62
Modelled: 62
Confidence: 21.1%
Identity: 19%
Fold: PAH2 domain
Superfamily: PAH2 domain
Family: PAH2 domain
Phyre2
| 4 |
|
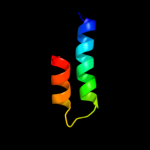
PDB 1vp8 chain A
Region: 85 - 92
Aligned: 8
Modelled: 8
Confidence: 18.7%
Identity: 63%
Fold: Pyruvate kinase C-terminal domain-like
Superfamily: PK C-terminal domain-like
Family: MTH1675-like
Phyre2
| 5 |
|
PDB 1t57 chain A
Region: 85 - 92
Aligned: 8
Modelled: 8
Confidence: 17.8%
Identity: 75%
Fold: Pyruvate kinase C-terminal domain-like
Superfamily: PK C-terminal domain-like
Family: MTH1675-like
Phyre2
| 6 |
|
PDB 3bl2 chain A domain 1
Region: 133 - 141
Aligned: 9
Modelled: 9
Confidence: 16.0%
Identity: 78%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 7 |
|
PDB 2rmr chain A
Region: 2 - 61
Aligned: 48
Modelled: 60
Confidence: 13.4%
Identity: 33%
PDB header:transcription
Chain: A: PDB Molecule:paired amphipathic helix protein sin3a;
PDBTitle: solution structure of msin3a pah1 domain
Phyre2
| 8 |
|
PDB 1utr chain A
Region: 48 - 65
Aligned: 18
Modelled: 18
Confidence: 12.5%
Identity: 28%
Fold: Uteroglobin-like
Superfamily: Uteroglobin-like
Family: Uteroglobin-like
Phyre2
| 9 |
|
PDB 1ccd chain A
Region: 48 - 65
Aligned: 18
Modelled: 18
Confidence: 12.3%
Identity: 28%
Fold: Uteroglobin-like
Superfamily: Uteroglobin-like
Family: Uteroglobin-like
Phyre2
| 10 |
|
PDB 1utg chain A
Region: 48 - 65
Aligned: 18
Modelled: 18
Confidence: 9.5%
Identity: 22%
Fold: Uteroglobin-like
Superfamily: Uteroglobin-like
Family: Uteroglobin-like
Phyre2
| 11 |
|
PDB 1n93 chain X
Region: 53 - 95
Aligned: 38
Modelled: 43
Confidence: 9.0%
Identity: 29%
Fold: P40 nucleoprotein
Superfamily: P40 nucleoprotein
Family: P40 nucleoprotein
Phyre2
| 12 |
|
PDB 1n93 chain X
Region: 53 - 95
Aligned: 38
Modelled: 43
Confidence: 9.0%
Identity: 29%
PDB header:viral protein
Chain: X: PDB Molecule:p40 nucleoprotein;
PDBTitle: crystal structure of the borna disease virus nucleoprotein
Phyre2
| 13 |
|
PDB 3qk9 chain B
Region: 12 - 62
Aligned: 50
Modelled: 51
Confidence: 7.4%
Identity: 14%
PDB header:protein transport
Chain: B: PDB Molecule:mitochondrial import inner membrane translocase subunit
PDBTitle: yeast tim44 c-terminal domain complexed with cymal-3
Phyre2
| 14 |
|
PDB 2fhd chain A
Region: 51 - 57
Aligned: 7
Modelled: 7
Confidence: 6.8%
Identity: 71%
PDB header:cell cycle
Chain: A: PDB Molecule:dna repair protein rhp9/crb2;
PDBTitle: crystal structure of crb2 tandem tudor domains
Phyre2
| 15 |
|
PDB 3bd0 chain D
Region: 17 - 57
Aligned: 41
Modelled: 41
Confidence: 6.6%
Identity: 20%
PDB header:peptide binding protein
Chain: D: PDB Molecule:protein memo1;
PDBTitle: crystal structure of memo, form ii
Phyre2
| 16 |
|
PDB 1mb4 chain A domain 1
Region: 70 - 104
Aligned: 35
Modelled: 35
Confidence: 6.2%
Identity: 26%
Fold: NAD(P)-binding Rossmann-fold domains
Superfamily: NAD(P)-binding Rossmann-fold domains
Family: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
Phyre2
| 17 |
|
PDB 1s28 chain A
Region: 114 - 140
Aligned: 27
Modelled: 27
Confidence: 6.1%
Identity: 26%
Fold: Secretion chaperone-like
Superfamily: Type III secretory system chaperone-like
Family: Type III secretory system chaperone
Phyre2
| 18 |
|
PDB 1nlx chain A
Region: 116 - 148
Aligned: 33
Modelled: 33
Confidence: 5.3%
Identity: 21%
Fold: Four-helical up-and-down bundle
Superfamily: Group V grass pollen allergen
Family: Group V grass pollen allergen
Phyre2