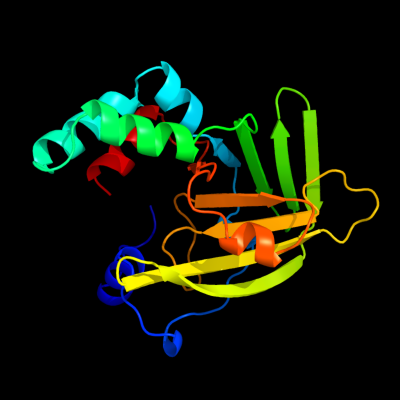
1 c1txlA_
100.0
100
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: metal-binding protein yoda;PDBTitle: crystal structure of metal-binding protein yoda from e.2 coli, pfam duf149
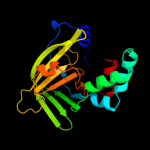
2 d1txla_
100.0
100
Fold: LipocalinsSuperfamily: LipocalinsFamily: Hypothetical protein YodA3 d1c8ba_
55.3
33
Fold: Phosphorylase/hydrolase-likeSuperfamily: HybD-likeFamily: Germination protease4 c2nvuB_
39.0
32
PDB header: protein turnover, ligaseChain: B: PDB Molecule: maltose binding protein/nedd8-activating enzymePDBTitle: structure of appbp1-uba3~nedd8-nedd8-mgatp-ubc12(c111a), a2 trapped ubiquitin-like protein activation complex
5 c2nogA_
29.7
20
PDB header: dna binding proteinChain: A: PDB Molecule: iswi protein;PDBTitle: sant domain structure of xenopus remodeling factor iswi
6 d1nkga3
27.3
12
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Rhamnogalacturonase B, RhgB, N-terminal domain7 c2kkmA_
27.2
29
PDB header: translationChain: A: PDB Molecule: translation machinery-associated protein 16;PDBTitle: solution nmr structure of yeast protein yor252w [residues2 38-178]: northeast structural genomics consortium target3 yt654
8 d1gjja1
24.0
19
Fold: LEM/SAP HeH motifSuperfamily: LEM domainFamily: LEM domain9 d1h9ea_
22.0
19
Fold: LEM/SAP HeH motifSuperfamily: LEM domainFamily: LEM domain10 d1yovb1
17.8
35
Fold: Activating enzymes of the ubiquitin-like proteinsSuperfamily: Activating enzymes of the ubiquitin-like proteinsFamily: Ubiquitin activating enzymes (UBA)11 c1x41A_
16.4
27
PDB header: transcriptionChain: A: PDB Molecule: transcriptional adaptor 2-like, isoform b;PDBTitle: solution structure of the myb-like dna binding domain of2 human transcriptional adaptor 2-like, isoform b
12 c3gznB_
15.1
35
PDB header: protein binding/ligaseChain: B: PDB Molecule: nedd8-activating enzyme e1 catalytic subunit;PDBTitle: structure of nedd8-activating enzyme in complex with nedd82 and mln4924
13 c2l5qA_
12.4
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of bvu_3817 from bacteroides vulgatus,2 northeast structural genomics consortium target bvr159
14 d1jlxa1
12.3
10
Fold: beta-TrefoilSuperfamily: AgglutininFamily: Agglutinin15 d1izna_
11.1
25
Fold: Subunits of heterodimeric actin filament capping protein CapzSuperfamily: Subunits of heterodimeric actin filament capping protein CapzFamily: Capz alpha-1 subunit16 c3lggA_
10.5
11
PDB header: hydrolaseChain: A: PDB Molecule: adenosine deaminase cecr1;PDBTitle: crystal structure of human adenosine deaminase growth factor,2 adenosine deaminase type 2 (ada2) complexed with transition state3 analogue, coformycin
17 d1q8ca_
10.2
19
Fold: NusB-likeSuperfamily: NusB-likeFamily: Hypothetical protein MG02718 d1w5ra1
10.2
15
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Arylamine N-acetyltransferase19 d3deoa1
9.4
23
Fold: SH3-like barrelSuperfamily: Chromo domain-likeFamily: Chromo domain20 d1cmca_
9.3
33
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Met repressor, MetJ (MetR)21 d1dixa_
not modelled
9.2
25
Fold: Ribonuclease Rh-likeSuperfamily: Ribonuclease Rh-likeFamily: Ribonuclease Rh-like22 d1ofcx1
not modelled
9.0
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Myb/SANT domain23 c2x0lB_
not modelled
8.6
19
PDB header: transcriptionChain: B: PDB Molecule: rest corepressor 1;PDBTitle: crystal structure of a neuro-specific splicing variant of2 human histone lysine demethylase lsd1.
24 c2iw5B_
not modelled
8.6
19
PDB header: oxidoreductase/transcription regulatorChain: B: PDB Molecule: rest corepressor 1;PDBTitle: structural basis for corest-dependent demethylation of2 nucleosomes by the human lsd1 histone demethylase
25 c2xajB_
not modelled
8.6
19
PDB header: transcriptionChain: B: PDB Molecule: rest corepressor 1;PDBTitle: crystal structure of lsd1-corest in complex with (-)-trans-2 2-phenylcyclopropyl-1-amine
26 c2v1dB_
not modelled
8.6
19
PDB header: oxidoreductase/repressorChain: B: PDB Molecule: rest corepressor 1;PDBTitle: structural basis of lsd1-corest selectivity in histone h32 recognition
27 d1xc5a1
not modelled
8.6
30
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Myb/SANT domain28 d2cqqa1
not modelled
8.6
7
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Myb/SANT domain29 d1mdwa_
not modelled
8.4
16
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Calpain large subunit, catalytic domain (domain II)30 d1a4ma_
not modelled
8.1
15
Fold: TIM beta/alpha-barrelSuperfamily: Metallo-dependent hydrolasesFamily: Adenosine/AMP deaminase31 d1x41a1
not modelled
8.1
33
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain32 d2ijra1
not modelled
8.1
19
Fold: Api92-likeSuperfamily: Api92-likeFamily: Api92-like33 d1vmha_
not modelled
7.8
57
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like34 d1j0ha1
not modelled
7.2
17
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes35 d1vmfa_
not modelled
7.1
71
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like36 c2kmuA_
not modelled
6.9
18
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent dna helicase q4;PDBTitle: recql4 amino-terminal domain
37 d1vmja_
not modelled
6.8
43
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like38 c2p6cB_
not modelled
6.7
43
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: aq_2013 protein;PDBTitle: crystal structure of hypothetical protein aq_2013 from aquifex2 aeolicus vf5.
39 c3oiqB_
not modelled
6.7
44
PDB header: protein bindingChain: B: PDB Molecule: dna polymerase alpha catalytic subunit a;PDBTitle: crystal structure of yeast telomere protein cdc13 ob1 and the2 catalytic subunit of dna polymerase alpha pol1
40 d1gvia1
not modelled
6.7
13
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes41 c3efcA_
not modelled
6.6
13
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein assembly factor yaet;PDBTitle: crystal structure of yaet periplasmic domain
42 c2cu5C_
not modelled
6.5
57
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: conserved hypothetical protein tt1486;PDBTitle: crystal structure of the conserved hypothetical protein tt1486 from2 thermus thermophilus hb8
43 c2vzkD_
not modelled
6.3
12
PDB header: transferaseChain: D: PDB Molecule: glutamate n-acetyltransferase 2 beta chain;PDBTitle: structure of the acyl-enzyme complex of an n-terminal2 nucleophile (ntn) hydrolase, oat2
44 d2ok5a4
not modelled
6.2
16
Fold: Complement control module/SCR domainSuperfamily: Complement control module/SCR domainFamily: Complement control module/SCR domain45 c1v91A_
not modelled
6.2
17
PDB header: toxinChain: A: PDB Molecule: delta-palutoxin it2;PDBTitle: solution structure of insectidal toxin delta-paluit2-nh2
46 d2fefa1
not modelled
6.1
32
Fold: Bromodomain-likeSuperfamily: PA2201 N-terminal domain-likeFamily: PA2201 N-terminal domain-like47 c3fljA_
not modelled
5.9
0
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein conserved in bacteria with aPDBTitle: crystal structure of uncharacterized protein conserved in bacteria2 with a cystatin-like fold (yp_168589.1) from silicibacter pomeroyi3 dss-3 at 2.00 a resolution
48 d2soba_
not modelled
5.9
11
Fold: OB-foldSuperfamily: Staphylococcal nucleaseFamily: Staphylococcal nuclease49 c3lnbA_
not modelled
5.8
6
PDB header: transferaseChain: A: PDB Molecule: n-acetyltransferase family protein;PDBTitle: crystal structure analysis of arylamine n-acetyltransferase c from2 bacillus anthracis
50 d1vpha_
not modelled
5.8
43
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like51 c1vraB_
not modelled
5.8
4
PDB header: transferaseChain: B: PDB Molecule: arginine biosynthesis bifunctional protein argj;PDBTitle: crystal structure of arginine biosynthesis bifunctional protein argj2 (10175521) from bacillus halodurans at 2.00 a resolution
52 d1xcra1
not modelled
5.8
31
Fold: AF0104/ALDC/Ptd012-likeSuperfamily: AF0104/ALDC/Ptd012-likeFamily: PTD012-like53 c1ve0A_
not modelled
5.7
43
PDB header: metal binding proteinChain: A: PDB Molecule: hypothetical protein (st2072);PDBTitle: crystal structure of uncharacterized protein st2072 from sulfolobus2 tokodaii
54 c3it4B_
not modelled
5.5
12
PDB header: transferaseChain: B: PDB Molecule: arginine biosynthesis bifunctional protein argjPDBTitle: the crystal structure of ornithine acetyltransferase from2 mycobacterium tuberculosis (rv1653) at 1.7 a
55 c3acgA_
not modelled
5.5
12
PDB header: hydrolaseChain: A: PDB Molecule: beta-1,4-endoglucanase;PDBTitle: crystal structure of carbohydrate-binding module family 282 from clostridium josui cel5a in complex with cellobiose
56 c2elkA_
not modelled
5.4
33
PDB header: transcriptionChain: A: PDB Molecule: spcc24b10.08c protein;PDBTitle: solution structure of the sant domain of fission yeast2 spcc24b10.08c protein
57 c2p6hB_
not modelled
5.3
43
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of hypothetical protein ape1520 from aeropyrum2 pernix k1
58 d2v94a1
not modelled
5.2
13
Fold: Ribosomal proteins S24e, L23 and L15eSuperfamily: Ribosomal proteins S24e, L23 and L15eFamily: Ribosomal protein S24e