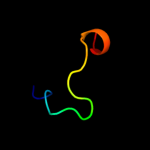
1 d1ig4a_
50.9
32
Fold: DNA-binding domainSuperfamily: DNA-binding domainFamily: Methyl-CpG-binding domain, MBD2 c2a2bA_
49.6
41
PDB header: antibioticChain: A: PDB Molecule: bacteriocin curvacin a;PDBTitle: curvacin a
3 c2q3eH_
16.6
29
PDB header: oxidoreductaseChain: H: PDB Molecule: udp-glucose 6-dehydrogenase;PDBTitle: structure of human udp-glucose dehydrogenase complexed with nadh and2 udp-glucose
4 d1flca1
11.7
45
Fold: Viral protein domainSuperfamily: Viral protein domainFamily: Hemagglutinin domain of haemagglutinin-esterase-fusion glycoprotein HEF15 d1ua7a1
11.0
10
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain6 c3ebkA_
9.9
22
PDB header: allergenChain: A: PDB Molecule: allergen bla g 4;PDBTitle: crystal structure of major allergens, bla g 4 from2 cockroaches
7 c2yqpA_
8.5
44
PDB header: gene regulation, hydrolaseChain: A: PDB Molecule: probable atp-dependent rna helicase ddx59;PDBTitle: solution structure of the zf-hit domain in dead (asp-glu-2 ala-asp) box polypeptide 59
8 d1q90r_
8.5
33
Fold: Single transmembrane helixSuperfamily: ISP transmembrane anchorFamily: ISP transmembrane anchor9 c1q90R_
8.5
33
PDB header: photosynthesisChain: R: PDB Molecule: cytochrome b6-f complex iron-sulfur subunit;PDBTitle: structure of the cytochrome b6f (plastohydroquinone : plastocyanin2 oxidoreductase) from chlamydomonas reinhardtii
10 c3plnA_
8.1
25
PDB header: oxidoreductaseChain: A: PDB Molecule: udp-glucose 6-dehydrogenase;PDBTitle: crystal structure of klebsiella pneumoniae udp-glucose 6-dehydrogenase2 complexed with udp-glucose
11 d1qkia2
7.6
45
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Glucose 6-phosphate dehydrogenase-like12 c3ggpA_
7.5
19
PDB header: oxidoreductaseChain: A: PDB Molecule: prephenate dehydrogenase;PDBTitle: crystal structure of prephenate dehydrogenase from a. aeolicus in2 complex with hydroxyphenyl propionate and nad+
13 d1hx0a1
7.1
40
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain14 c3prjB_
7.0
29
PDB header: oxidoreductaseChain: B: PDB Molecule: udp-glucose 6-dehydrogenase;PDBTitle: role of packing defects in the evolution of allostery and induced fit2 in human udp-glucose dehydrogenase.
15 d1g94a1
6.6
40
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain16 d1jaea1
6.6
40
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain17 c2bhlB_
6.6
45
PDB header: oxidoreductase (choh(d)-nadp)Chain: B: PDB Molecule: glucose-6-phosphate 1-dehydrogenase;PDBTitle: x-ray structure of human glucose-6-phosphate dehydrogenase2 (deletion variant) complexed with glucose-6-phosphate
18 c1e0fI_
6.5
37
PDB header: coagulation/crystal structure/heparin-bChain: I: PDB Molecule: haemadin;PDBTitle: crystal structure of the human alpha-thrombin-haemadin2 complex: an exosite ii-binding inhibitor
19 d1e0fi_
6.5
37
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Leech antihemostatic proteinsFamily: Hirudin-like20 d3dhpa1
6.3
40
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain21 d1t27a_
not modelled
6.2
16
Fold: TBP-likeSuperfamily: Bet v1-likeFamily: Phoshatidylinositol transfer protein, PITP22 d1r0va3
not modelled
6.1
29
Fold: MutS N-terminal domain-likeSuperfamily: tRNA-intron endonuclease N-terminal domain-likeFamily: tRNA-intron endonuclease N-terminal domain-like23 d1rmka_
not modelled
6.0
45
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Conotoxin24 d1h9aa1
not modelled
6.0
33
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain25 c2l3bA_
not modelled
5.9
29
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein found in conjugate transposon;PDBTitle: solution nmr structure of the bt_0084 lipoprotein from bacteroides2 thetaiotaomicron, northeast structural genomics consortium target3 btr376
26 d2a1la1
not modelled
5.9
20
Fold: TBP-likeSuperfamily: Bet v1-likeFamily: Phoshatidylinositol transfer protein, PITP27 c2l7qA_
not modelled
5.9
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein found in conjugate transposon;PDBTitle: solution nmr structure of conjugate transposon protein bvu_1572(27-2 141) from bacteroides vulgatus, northeast structural genomics3 consortium target bvr155
28 c1e0fJ_
not modelled
5.8
37
PDB header: coagulation/crystal structure/heparin-bChain: J: PDB Molecule: haemadin;PDBTitle: crystal structure of the human alpha-thrombin-haemadin2 complex: an exosite ii-binding inhibitor
29 c1qkiE_
not modelled
5.5
45
PDB header: oxidoreductaseChain: E: PDB Molecule: glucose-6-phosphate 1-dehydrogenase;PDBTitle: x-ray structure of human glucose 6-phosphate dehydrogenase2 (variant canton r459l) complexed with structural nadp+