| 1 | c3fmxX_ |
|
 |
100.0 |
78 |
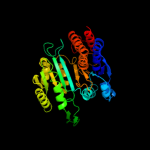
PDB header:oxidoreductase
Chain: X: PDB Molecule:tartrate dehydrogenase/decarboxylase;
PDBTitle: crystal structure of tartrate dehydrogenase from pseudomonas2 putida complexed with nadh
|
| 2 | c2d1cB_ |
|
 |
100.0 |
35 |
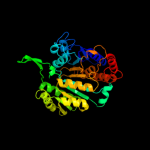
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: crystal structure of tt0538 protein from thermus thermophilus hb8
|
| 3 | d1cm7a_ |
|
 |
100.0 |
38 |
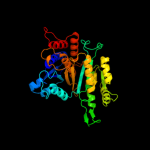
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 4 | d1pb1a_ |
|
 |
100.0 |
29 |
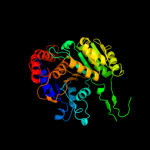
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 5 | d1v53a1 |
|
 |
100.0 |
36 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 6 | d1cnza_ |
|
 |
100.0 |
37 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 7 | d1a05a_ |
|
 |
100.0 |
34 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 8 | c3r8wC_ |
|
 |
100.0 |
36 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:3-isopropylmalate dehydrogenase 2, chloroplastic;
PDBTitle: structure of 3-isopropylmalate dehydrogenase isoform 2 from2 arabidopsis thaliana at 2.2 angstrom resolution
|
| 9 | c2d4vD_ |
|
 |
100.0 |
29 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: crystal structure of nad dependent isocitrate dehydrogenase2 from acidithiobacillus thiooxidans
|
| 10 | d1g2ua_ |
|
 |
100.0 |
36 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 11 | d1xaca_ |
|
 |
100.0 |
36 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 12 | d1hqsa_ |
|
 |
100.0 |
31 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 13 | d1vlca_ |
|
 |
100.0 |
36 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 14 | c2e0cA_ |
|
 |
100.0 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:409aa long hypothetical nadp-dependent isocitrate
PDBTitle: crystal structure of isocitrate dehydrogenase from sulfolobus tokodaii2 strain7 at 2.0 a resolution
|
| 15 | c3u1hA_ |
|
 |
100.0 |
40 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-isopropylmalate dehydrogenase;
PDBTitle: crystal structure of ipmdh from the last common ancestor of bacillus
|
| 16 | d1wpwa_ |
|
 |
100.0 |
34 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 17 | c1x0lB_ |
|
 |
100.0 |
42 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:homoisocitrate dehydrogenase;
PDBTitle: crystal structure of tetrameric homoisocitrate dehydrogenase from an2 extreme thermophile, thermus thermophilus
|
| 18 | d1w0da_ |
|
 |
100.0 |
41 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 19 | c3blxL_ |
|
 |
100.0 |
33 |
PDB header:oxidoreductase
Chain: L: PDB Molecule:isocitrate dehydrogenase [nad] subunit 2;
PDBTitle: yeast isocitrate dehydrogenase (apo form)
|
| 20 | c1tyoA_ |
|
 |
100.0 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: isocitrate dehydrogenase from the hyperthermophile aeropyrum pernix in2 complex with etheno-nadp
|
| 21 | c3blxM_ |
|
not modelled |
100.0 |
31 |
PDB header:oxidoreductase
Chain: M: PDB Molecule:isocitrate dehydrogenase [nad] subunit 1;
PDBTitle: yeast isocitrate dehydrogenase (apo form)
|
| 22 | c1zorB_ |
|
not modelled |
100.0 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: isocitrate dehydrogenase from the hyperthermophile thermotoga maritima
|
| 23 | c2uxqB_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase native;
PDBTitle: isocitrate dehydrogenase from the psychrophilic bacterium2 desulfotalea psychrophila: biochemical properties and3 crystal structure analysis
|
| 24 | d1lwda_ |
|
not modelled |
100.0 |
18 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 25 | c3us8A_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase [nadp];
PDBTitle: crystal structure of an isocitrate dehydrogenase from sinorhizobium2 meliloti 1021
|
| 26 | d1t0la_ |
|
not modelled |
100.0 |
17 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 27 | c2qfyE_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:isocitrate dehydrogenase [nadp];
PDBTitle: crystal structure of saccharomyces cerevesiae mitochondrial nadp(+)-2 dependent isocitrate dehydrogenase in complex with a-ketoglutarate
|
| 28 | c2iv0A_ |
|
not modelled |
100.0 |
32 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: thermal stability of isocitrate dehydrogenase from2 archaeoglobus fulgidus studied by crystal structure3 analysis and engineering of chimers
|
| 29 | c2hi1A_ |
|
not modelled |
99.0 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:4-hydroxythreonine-4-phosphate dehydrogenase 2;
PDBTitle: the structure of a putative 4-hydroxythreonine-4-phosphate2 dehydrogenase from salmonella typhimurium.
|
| 30 | d1ptma_ |
|
not modelled |
99.0 |
16 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:PdxA-like |
| 31 | d1r8ka_ |
|
not modelled |
98.8 |
17 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:PdxA-like |
| 32 | c1yxoB_ |
|
not modelled |
98.5 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-hydroxythreonine-4-phosphate dehydrogenase 1;
PDBTitle: crystal structure of pyridoxal phosphate biosynthetic protein pdxa2 pa0593
|
| 33 | d1itwa_ |
|
not modelled |
98.1 |
18 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Monomeric isocitrate dehydrogenase |
| 34 | c2b0tA_ |
|
not modelled |
98.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadp isocitrate dehydrogenase;
PDBTitle: structure of monomeric nadp isocitrate dehydrogenase
|
| 35 | c2q62A_ |
|
not modelled |
77.0 |
10 |
PDB header:flavoprotein
Chain: A: PDB Molecule:arsh;
PDBTitle: crystal structure of arsh from sinorhizobium meliloti
|
| 36 | d2fpoa1 |
|
not modelled |
66.7 |
10 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:YhhF-like |
| 37 | c1p84E_ |
|
not modelled |
57.1 |
32 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur
PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
|
| 38 | d2onsa1 |
|
not modelled |
53.1 |
13 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 39 | c3cfxA_ |
|
not modelled |
53.0 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:upf0100 protein ma_0280;
PDBTitle: crystal structure of m. acetivorans periplasmic binding protein2 moda/wtpa with bound tungstate
|
| 40 | c2fyuE_ |
|
not modelled |
51.8 |
32 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit,
PDBTitle: crystal structure of bovine heart mitochondrial bc1 with jg1442 inhibitor
|
| 41 | c3s40C_ |
|
not modelled |
49.1 |
11 |
PDB header:transferase
Chain: C: PDB Molecule:diacylglycerol kinase;
PDBTitle: the crystal structure of a diacylglycerol kinases from bacillus2 anthracis str. sterne
|
| 42 | c3cfzA_ |
|
not modelled |
48.2 |
9 |
PDB header:transport protein
Chain: A: PDB Molecule:upf0100 protein mj1186;
PDBTitle: crystal structure of m. jannaschii periplasmic binding2 protein moda/wtpa with bound tungstate
|
| 43 | d3thia_ |
|
not modelled |
44.0 |
18 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 44 | d1riea_ |
|
not modelled |
41.0 |
32 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 45 | c3k6wA_ |
|
not modelled |
40.4 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:solute-binding protein ma_0280;
PDBTitle: apo and ligand bound structures of moda from the archaeon2 methanosarcina acetivorans
|
| 46 | c2fynO_ |
|
not modelled |
39.6 |
29 |
PDB header:oxidoreductase
Chain: O: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur
PDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
|
| 47 | c1orhA_ |
|
not modelled |
37.3 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:protein arginine n-methyltransferase 1;
PDBTitle: structure of the predominant protein arginine methyltransferase prmt1
|
| 48 | d1oria_ |
|
not modelled |
36.4 |
10 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Arginine methyltransferase |
| 49 | d3cx5e1 |
|
not modelled |
32.1 |
32 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 50 | c2e76D_ |
|
not modelled |
28.5 |
38 |
PDB header:photosynthesis
Chain: D: PDB Molecule:cytochrome b6-f complex iron-sulfur subunit;
PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
|
| 51 | c1twyG_ |
|
not modelled |
26.8 |
16 |
PDB header:structural genomics, unknown function
Chain: G: PDB Molecule:abc transporter, periplasmic substrate-binding protein;
PDBTitle: crystal structure of an abc-type phosphate transport receptor from2 vibrio cholerae
|
| 52 | d1twya_ |
|
not modelled |
26.8 |
16 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 53 | c3fzgA_ |
|
not modelled |
25.7 |
6 |
PDB header:transferase
Chain: A: PDB Molecule:16s rrna methylase;
PDBTitle: structure of the 16s rrna methylase arma
|
| 54 | c3grzA_ |
|
not modelled |
24.6 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:ribosomal protein l11 methyltransferase;
PDBTitle: crystal structure of ribosomal protein l11 methylase from2 lactobacillus delbrueckii subsp. bulgaricus
|
| 55 | c2nvgA_ |
|
not modelled |
24.1 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit;
PDBTitle: soluble domain of rieske iron sulfur protein.
|
| 56 | c3qd7X_ |
|
not modelled |
23.5 |
5 |
PDB header:hydrolase
Chain: X: PDB Molecule:uncharacterized protein ydal;
PDBTitle: crystal structure of ydal, a stand-alone small muts-related protein2 from escherichia coli
|
| 57 | d1ydga_ |
|
not modelled |
23.1 |
15 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:WrbA-like |
| 58 | d2fyta1 |
|
not modelled |
22.9 |
12 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Arginine methyltransferase |
| 59 | c2huzB_ |
|
not modelled |
17.2 |
13 |
PDB header:structural genomics, transferase
Chain: B: PDB Molecule:glucosamine 6-phosphate n-acetyltransferase;
PDBTitle: crystal structure of gnpnat1
|
| 60 | d2fcaa1 |
|
not modelled |
16.2 |
10 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:TrmB-like |
| 61 | c3lwzC_ |
|
not modelled |
16.1 |
24 |
PDB header:lyase
Chain: C: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: 1.65 angstrom resolution crystal structure of type ii 3-2 dehydroquinate dehydratase (aroq) from yersinia pestis
|
| 62 | d2z3va1 |
|
not modelled |
16.0 |
12 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 63 | d1t0ia_ |
|
not modelled |
15.8 |
19 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:NADPH-dependent FMN reductase |
| 64 | d1yzha1 |
|
not modelled |
15.5 |
6 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:TrmB-like |
| 65 | d1g6q1_ |
|
not modelled |
15.3 |
10 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Arginine methyltransferase |
| 66 | c3ksmA_ |
|
not modelled |
15.2 |
13 |
PDB header:transport protein
Chain: A: PDB Molecule:abc-type sugar transport system, periplasmic component;
PDBTitle: crystal structure of abc-type sugar transport system, periplasmic2 component from hahella chejuensis
|
| 67 | c2pebB_ |
|
not modelled |
15.1 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative dioxygenase;
PDBTitle: crystal structure of a putative dioxygenase (npun_f1925) from nostoc2 punctiforme pcc 73102 at 1.46 a resolution
|
| 68 | c2qhxB_ |
|
not modelled |
15.0 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:pteridine reductase 1;
PDBTitle: structure of pteridine reductase from leishmania major2 complexed with a ligand
|
| 69 | d1n1jb_ |
|
not modelled |
14.9 |
11 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:TBP-associated factors, TAFs |
| 70 | d1tzyb_ |
|
not modelled |
14.8 |
38 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 71 | c3fg9B_ |
|
not modelled |
14.6 |
18 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:protein of universal stress protein uspa family;
PDBTitle: the crystal structure of an universal stress protein uspa2 family protein from lactobacillus plantarum wcfs1
|
| 72 | d1ofda1 |
|
not modelled |
14.6 |
19 |
Fold:Single-stranded right-handed beta-helix
Superfamily:Alpha subunit of glutamate synthase, C-terminal domain
Family:Alpha subunit of glutamate synthase, C-terminal domain |
| 73 | d1hioa_ |
|
not modelled |
13.4 |
11 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 74 | d1cyxa_ |
|
not modelled |
13.3 |
21 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Periplasmic domain of cytochrome c oxidase subunit II |
| 75 | c1cyxA_ |
|
not modelled |
13.3 |
21 |
PDB header:electron transport
Chain: A: PDB Molecule:cyoa;
PDBTitle: quinol oxidase (periplasmic fragment of subunit ii with2 engineered cu-a binding site)(cyoa)
|
| 76 | d1r5ja_ |
|
not modelled |
13.2 |
17 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Phosphotransacetylase |
| 77 | c2uvgA_ |
|
not modelled |
13.2 |
14 |
PDB header:sugar-binding protein
Chain: A: PDB Molecule:abc type periplasmic sugar-binding protein;
PDBTitle: structure of a periplasmic oligogalacturonide binding2 protein from yersinia enterocolitica
|
| 78 | c1f66C_ |
|
not modelled |
12.6 |
11 |
PDB header:structural protein/dna
Chain: C: PDB Molecule:histone h2a.z;
PDBTitle: 2.6 a crystal structure of a nucleosome core particle2 containing the variant histone h2a.z
|
| 79 | d1f66c_ |
|
not modelled |
12.6 |
11 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 80 | c2hxrA_ |
|
not modelled |
11.9 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:hth-type transcriptional regulator cynr;
PDBTitle: structure of the ligand binding domain of e. coli cynr, a2 transcriptional regulator controlling cyanate metabolism
|
| 81 | c3er6D_ |
|
not modelled |
11.8 |
17 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:putative transcriptional regulator protein;
PDBTitle: crystal structure of a putative transcriptional regulator2 protein from vibrio parahaemolyticus
|
| 82 | d1id3d_ |
|
not modelled |
11.8 |
29 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 83 | d1n1ja_ |
|
not modelled |
11.6 |
16 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:TBP-associated factors, TAFs |
| 84 | c3b3jA_ |
|
not modelled |
11.6 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:histone-arginine methyltransferase carm1;
PDBTitle: the 2.55 a crystal structure of the apo catalytic domain of2 coactivator-associated arginine methyl transferase i(carm1:28-507,3 residues 28-146 and 479-507 not ordered)
|
| 85 | d1i27a_ |
|
not modelled |
11.5 |
22 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:C-terminal domain of the rap74 subunit of TFIIF |
| 86 | d2jssa1 |
|
not modelled |
11.5 |
14 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 87 | c2f8nK_ |
|
not modelled |
11.4 |
11 |
PDB header:structural protein/dna
Chain: K: PDB Molecule:histone h2a type 1;
PDBTitle: 2.9 angstrom x-ray structure of hybrid macroh2a nucleosomes
|
| 88 | c3c6kC_ |
|
not modelled |
11.2 |
10 |
PDB header:transferase
Chain: C: PDB Molecule:spermine synthase;
PDBTitle: crystal structure of human spermine synthase in complex2 with spermidine and 5-methylthioadenosine
|
| 89 | c2qv7A_ |
|
not modelled |
11.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:diacylglycerol kinase dgkb;
PDBTitle: crystal structure of diacylglycerol kinase dgkb in complex with adp2 and mg
|
| 90 | d1fftb1 |
|
not modelled |
10.8 |
21 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Periplasmic domain of cytochrome c oxidase subunit II |
| 91 | d1e7wa_ |
|
not modelled |
10.7 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 92 | d1i7na1 |
|
not modelled |
10.6 |
25 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:Synapsin domain |
| 93 | d1jq4a_ |
|
not modelled |
10.4 |
23 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 94 | d1p3mh_ |
|
not modelled |
10.4 |
38 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 95 | c2zqeA_ |
|
not modelled |
10.3 |
11 |
PDB header:dna binding protein
Chain: A: PDB Molecule:muts2 protein;
PDBTitle: crystal structure of the smr domain of thermus thermophilus muts2
|
| 96 | d1q77a_ |
|
not modelled |
10.3 |
13 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 97 | d1ku5a_ |
|
not modelled |
10.1 |
33 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Archaeal histone |
| 98 | d1hiob_ |
|
not modelled |
10.0 |
38 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 99 | d1tzya_ |
|
not modelled |
10.0 |
11 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |

