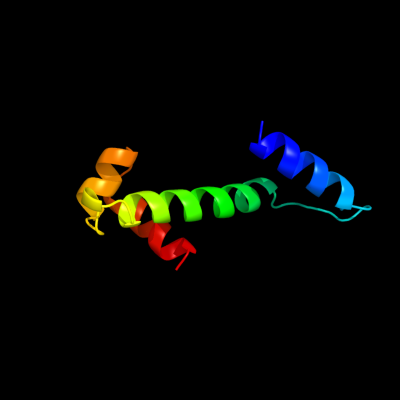
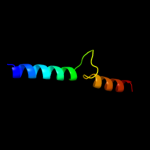
1 d1fipa_
20.8
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like2 d2ns0a1
20.5
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RHA1 ro06458-like3 c3k6qB_
19.0
12
PDB header: ligand binding proteinChain: B: PDB Molecule: putative ligand binding protein;PDBTitle: crystal structure of an antitoxin part of a putative toxin/antitoxin2 system (swol_0700) from syntrophomonas wolfei subsp. wolfei at 1.80 a3 resolution
4 d1w36c2
17.7
9
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain5 d1etxa_
14.3
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like6 d1nj1a2
14.0
29
Fold: IF3-likeSuperfamily: C-terminal domain of ProRSFamily: C-terminal domain of ProRS7 d2ak3a2
11.0
26
Fold: Rubredoxin-likeSuperfamily: Microbial and mitochondrial ADK, insert "zinc finger" domainFamily: Microbial and mitochondrial ADK, insert "zinc finger" domain8 d1e4va2
10.4
30
Fold: Rubredoxin-likeSuperfamily: Microbial and mitochondrial ADK, insert "zinc finger" domainFamily: Microbial and mitochondrial ADK, insert "zinc finger" domain9 d1p3ja2
10.3
30
Fold: Rubredoxin-likeSuperfamily: Microbial and mitochondrial ADK, insert "zinc finger" domainFamily: Microbial and mitochondrial ADK, insert "zinc finger" domain10 d1t6sa1
10.3
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ScpB/YpuH-like11 d1zina2
10.2
35
Fold: Rubredoxin-likeSuperfamily: Microbial and mitochondrial ADK, insert "zinc finger" domainFamily: Microbial and mitochondrial ADK, insert "zinc finger" domain12 d1s3ga2
9.4
35
Fold: Rubredoxin-likeSuperfamily: Microbial and mitochondrial ADK, insert "zinc finger" domainFamily: Microbial and mitochondrial ADK, insert "zinc finger" domain13 d1akya2
8.9
22
Fold: Rubredoxin-likeSuperfamily: Microbial and mitochondrial ADK, insert "zinc finger" domainFamily: Microbial and mitochondrial ADK, insert "zinc finger" domain14 d1poia_
7.6
8
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: CoA transferase alpha subunit-like15 d1g2ha_
7.5
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like16 d1xx6a1
7.4
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Type II thymidine kinase17 c1qhhB_
6.9
13
PDB header: hydrolaseChain: B: PDB Molecule: protein (pcra (subunit));PDBTitle: structure of dna helicase with adpnp
18 c2qfaC_
5.9
17
PDB header: cell cycle/cell cycle/cell cycleChain: C: PDB Molecule: inner centromere protein;PDBTitle: crystal structure of a survivin-borealin-incenp core complex
19 c3ndhA_
5.9
22
PDB header: hydrolase/dnaChain: A: PDB Molecule: restriction endonuclease thai;PDBTitle: restriction endonuclease in complex with substrate dna
20 c2wdtA_
5.8
9
PDB header: hydrolase/protein bindingChain: A: PDB Molecule: ubiquitin carboxyl-terminal hydrolase l3;PDBTitle: crystal structure of plasmodium falciparum uchl3 in complex2 with the suicide inhibitor ubvme
21 d2f7fa2
not modelled
5.7
50
Fold: alpha/beta-HammerheadSuperfamily: Nicotinate/Quinolinate PRTase N-terminal domain-likeFamily: NadC N-terminal domain-like22 d1nbwa1
not modelled
5.4
22
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Swiveling domain of dehydratase reactivase alpha subunitFamily: Swiveling domain of dehydratase reactivase alpha subunit23 c3e2iA_
not modelled
5.3
11
PDB header: transferaseChain: A: PDB Molecule: thymidine kinase;PDBTitle: crystal structure of thymidine kinase from s. aureus
24 d1etob_
not modelled
5.3
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like25 c1nj2A_
not modelled
5.2
29
PDB header: ligaseChain: A: PDB Molecule: proline-trna synthetase;PDBTitle: crystal structure of prolyl-trna synthetase from methanothermobacter2 thermautotrophicus