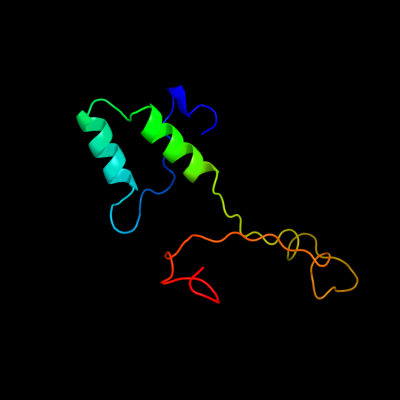
1 c4a1oB_
26.1
19
PDB header: transferase-hydrolaseChain: B: PDB Molecule: bifunctional purine biosynthesis protein purh;PDBTitle: crystal structure of mycobacterium tuberculosis purh complexed with2 aicar and a novel nucleotide cfair, at 2.48 a resolution.
2 d2g39a1
24.3
21
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: CoA transferase alpha subunit-like3 c2no8A_
13.6
31
PDB header: immune systemChain: A: PDB Molecule: colicin-e2 immunity protein;PDBTitle: nmr structure analysis of the colicin immuntiy protein im2
4 c2l95A_
13.5
8
PDB header: hydrolaseChain: A: PDB Molecule: crammer;PDBTitle: solution structure of cytotoxic t-lymphocyte antigent-2(ctla protein),2 crammer at ph 6.0
5 c3bq7A_
12.3
29
PDB header: transferaseChain: A: PDB Molecule: diacylglycerol kinase delta;PDBTitle: sam domain of diacylglycerol kinase delta1 (e35g)
6 d1iv8a2
12.3
47
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain7 d1zo0a1
11.9
7
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: Ornithine decarboxylase antizyme-like8 d2csba4
11.8
33
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Topoisomerase V repeat domain9 c2xqoA_
11.6
38
PDB header: hydrolaseChain: A: PDB Molecule: cellulosome enzyme, dockerin type i;PDBTitle: ctcel124: a cellulase from clostridium thermocellum
10 c3hjeA_
11.1
47
PDB header: transferaseChain: A: PDB Molecule: 704aa long hypothetical glycosyltransferase;PDBTitle: crystal structure of sulfolobus tokodaii hypothetical2 maltooligosyl trehalose synthase
11 c1v85A_
10.3
11
PDB header: apoptosisChain: A: PDB Molecule: similar to ring finger protein 36;PDBTitle: sterile alpha motif (sam) domain of mouse bifunctional2 apoptosis regulator
12 c3es5A_
9.1
45
PDB header: virusChain: A: PDB Molecule: putative capsid protein;PDBTitle: crystal structure of partitivirus (psv-f)
13 d1gxha_
8.9
23
Fold: Acyl carrier protein-likeSuperfamily: Colicin E immunity proteinsFamily: Colicin E immunity proteins14 c3op0B_
8.7
21
PDB header: signaling protein/signaling protein reguChain: B: PDB Molecule: signal transduction protein cbl-c;PDBTitle: crystal structure of cbl-c (cbl-3) tkb domain in complex with egfr2 py1069 peptide
15 c2nogA_
8.4
21
PDB header: dna binding proteinChain: A: PDB Molecule: iswi protein;PDBTitle: sant domain structure of xenopus remodeling factor iswi
16 d2dlda2
8.3
24
Fold: Flavodoxin-likeSuperfamily: Formate/glycerate dehydrogenase catalytic domain-likeFamily: Formate/glycerate dehydrogenases, substrate-binding domain17 c3bs7A_
8.1
18
PDB header: signaling proteinChain: A: PDB Molecule: protein aveugle;PDBTitle: crystal structure of the sterile alpha motif (sam) domain2 of hyphen/aveugle
18 c1l6lK_
8.0
29
PDB header: lipid transportChain: K: PDB Molecule: apolipoprotein a-ii;PDBTitle: structures of apolipoprotein a-ii and a lipid surrogate2 complex provide insights into apolipoprotein-lipid3 interactions
19 d2vlqa1
7.4
23
Fold: Acyl carrier protein-likeSuperfamily: Colicin E immunity proteinsFamily: Colicin E immunity proteins20 d1j4aa2
7.4
31
Fold: Flavodoxin-likeSuperfamily: Formate/glycerate dehydrogenase catalytic domain-likeFamily: Formate/glycerate dehydrogenases, substrate-binding domain21 c1ofcX_
not modelled
7.4
29
PDB header: nuclear proteinChain: X: PDB Molecule: iswi protein;PDBTitle: nucleosome recognition module of iswi atpase
22 d2cqqa1
not modelled
7.3
27
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Myb/SANT domain23 d1f0la2
not modelled
7.3
29
Fold: ADP-ribosylationSuperfamily: ADP-ribosylationFamily: ADP-ribosylating toxins24 d2hqya2
not modelled
6.9
19
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: FemXAB nonribosomal peptidyltransferases25 c2wsfG_
not modelled
6.7
47
PDB header: photosynthesisChain: G: PDB Molecule: photosystem i reaction center subunit v,PDBTitle: improved model of plant photosystem i
26 c3f75P_
not modelled
6.7
38
PDB header: hydrolaseChain: P: PDB Molecule: cathepsin l propeptide;PDBTitle: activated toxoplasma gondii cathepsin l (tgcpl) in complex with its2 propeptide
27 c2wscK_
not modelled
6.5
26
PDB header: photosynthesisChain: K: PDB Molecule: photosystem i reaction center subunit psak,PDBTitle: improved model of plant photosystem i
28 d2fu5a1
not modelled
6.5
25
Fold: Mss4-likeSuperfamily: Mss4-likeFamily: RabGEF Mss429 d3buxb1
not modelled
6.2
24
Fold: EF Hand-likeSuperfamily: EF-handFamily: EF-hand modules in multidomain proteins30 d1ayia_
not modelled
6.2
23
Fold: Acyl carrier protein-likeSuperfamily: Colicin E immunity proteinsFamily: Colicin E immunity proteins31 d1wwva1
not modelled
6.1
14
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain32 c3qodB_
not modelled
6.1
14
PDB header: dna binding proteinChain: B: PDB Molecule: heterocyst differentiation protein;PDBTitle: crystal structure of heterocyst differentiation protein, hetr from2 fischerella mv11
33 d1dpsa_
not modelled
5.7
71
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin34 d1t92a_
not modelled
5.7
18
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pseudopilin35 c3cecA_
not modelled
5.5
57
PDB header: transcriptionChain: A: PDB Molecule: putative antidote protein of plasmid maintenance system;PDBTitle: crystal structure of a putative antidote protein of plasmid2 maintenance system (npun_f2943) from nostoc punctiforme pcc 73102 at3 1.60 a resolution
36 c2o01G_
not modelled
5.3
47
PDB header: photosynthesisChain: G: PDB Molecule: photosystem i reaction center subunit v,PDBTitle: the structure of a plant photosystem i supercomplex at 3.42 angstrom resolution