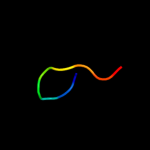
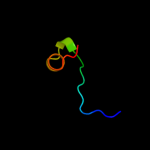
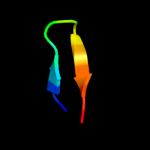
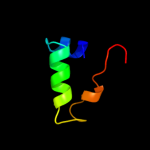
1 d1sg7a1
49.3
46
Fold: ChaB-likeSuperfamily: ChaB-likeFamily: ChaB-like2 c1sg7A_
49.3
46
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative cation transport regulator chab;PDBTitle: nmr solution structure of the putative cation transport2 regulator chab
3 d1lq1a_
45.0
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: Spo0A4 d1fc3a_
44.8
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: Spo0A5 d2ikba1
26.3
31
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: NMB1012-like6 c2l9yA_
22.3
13
PDB header: sugar binding proteinChain: A: PDB Molecule: cvnh-lysm lectin;PDBTitle: solution structure of the mocvnh-lysm module from the rice blast2 fungus magnaporthe oryzae protein (mgg_03307)
7 d2nr7a1
21.7
16
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: NMB1012-like8 d2jnaa1
18.0
15
Fold: Dodecin subunit-likeSuperfamily: YdgH-likeFamily: YdgH-like9 c2k9iB_
17.3
24
PDB header: dna binding proteinChain: B: PDB Molecule: plasmid prn1, complete sequence;PDBTitle: nmr structure of plasmid copy control protein orf56 from2 sulfolobus islandicus
10 c2cg4B_
17.1
23
PDB header: transcriptionChain: B: PDB Molecule: regulatory protein asnc;PDBTitle: structure of e.coli asnc
11 c2bw2A_
16.8
14
PDB header: signaling proteinChain: A: PDB Molecule: bypass of forespore c;PDBTitle: bofc from bacillus subtilis
12 d1ttya_
14.6
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain13 c1rk8C_
14.4
22
PDB header: translationChain: C: PDB Molecule: within the bgcn gene intron protein;PDBTitle: structure of the cytosolic protein pym bound to the mago-2 y14 core of the exon junction complex
14 d1rk8c_
14.4
22
Fold: WW domain-likeSuperfamily: Pym (Within the bgcn gene intron protein, WIBG), N-terminal domainFamily: Pym (Within the bgcn gene intron protein, WIBG), N-terminal domain15 c3lpeF_
14.4
25
PDB header: transferaseChain: F: PDB Molecule: dna-directed rna polymerase subunit e'';PDBTitle: crystal structure of spt4/5ngn heterodimer complex from methanococcus2 jannaschii
16 d1qqha_
13.8
27
Fold: E2 regulatory, transactivation domainSuperfamily: E2 regulatory, transactivation domainFamily: E2 regulatory, transactivation domain17 c1mgtA_
13.6
24
PDB header: transferaseChain: A: PDB Molecule: protein (o6-methylguanine-dna methyltransferase);PDBTitle: crystal structure of o6-methylguanine-dna methyltransferase from2 hyperthermophilic archaeon pyrococcus kodakaraensis strain kod1
18 d1gzsb_
12.5
17
Fold: SopE-like GEF domainSuperfamily: SopE-like GEF domainFamily: SopE-like GEF domain19 d2cg4a1
12.5
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain20 d2cyya1
11.8
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain21 c2ysaA_
not modelled
11.7
50
PDB header: metal binding proteinChain: A: PDB Molecule: retinoblastoma-binding protein 6;PDBTitle: solution structure of the zinc finger cchc domain from the2 human retinoblastoma-binding protein 6 (retinoblastoma-3 binding q protein 1, rbq-1)
22 d1tueb_
not modelled
11.6
27
Fold: E2 regulatory, transactivation domainSuperfamily: E2 regulatory, transactivation domainFamily: E2 regulatory, transactivation domain23 c2o8xA_
not modelled
11.6
22
PDB header: transcriptionChain: A: PDB Molecule: probable rna polymerase sigma-c factor;PDBTitle: crystal structure of the "-35 element" promoter recognition domain of2 mycobacterium tuberculosis sigc
24 d1ku7a_
not modelled
11.3
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain25 d1r6ea_
not modelled
11.3
19
Fold: SopE-like GEF domainSuperfamily: SopE-like GEF domainFamily: SopE-like GEF domain26 c1tueG_
not modelled
10.9
27
PDB header: replicationChain: G: PDB Molecule: regulatory protein e2;PDBTitle: the x-ray structure of the papillomavirus helicase in2 complex with its molecular matchmaker e2
27 c3frwF_
not modelled
10.5
16
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: putative trp repressor protein;PDBTitle: crystal structure of putative trpr protein from ruminococcus obeum
28 d2ilaa_
not modelled
10.4
27
Fold: beta-TrefoilSuperfamily: CytokineFamily: Interleukin-1 (IL-1)29 c2c5kP_
not modelled
9.8
21
PDB header: protein transportChain: P: PDB Molecule: vacuolar protein sorting protein 51;PDBTitle: n-terminal domain of tlg1 complexed with n-terminus of2 vps51
30 c1l0oC_
not modelled
9.6
12
PDB header: protein bindingChain: C: PDB Molecule: sigma factor;PDBTitle: crystal structure of the bacillus stearothermophilus anti-2 sigma factor spoiiab with the sporulation sigma factor3 sigmaf
31 d1l0oc_
not modelled
9.6
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain32 d1rp3a1
not modelled
9.6
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma3 domain33 c3korD_
not modelled
9.4
16
PDB header: transcriptionChain: D: PDB Molecule: possible trp repressor;PDBTitle: crystal structure of a putative trp repressor from staphylococcus2 aureus
34 c2dbbA_
not modelled
9.1
23
PDB header: transcriptional regulatorChain: A: PDB Molecule: putative hth-type transcriptional regulator ph0061;PDBTitle: crystal structure of ph0061
35 c2fgtA_
not modelled
8.8
38
PDB header: signaling proteinChain: A: PDB Molecule: two-component system yycf/yycg regulatoryPDBTitle: crystal structure of yych from bacillus subtilis
36 c2e1cA_
not modelled
8.6
14
PDB header: transcription/dnaChain: A: PDB Molecule: putative hth-type transcriptional regulator ph1519;PDBTitle: structure of putative hth-type transcriptional regulator ph1519/dna2 complex
37 c3h8dC_
not modelled
8.4
21
PDB header: motor protein/signaling proteinChain: C: PDB Molecule: myosin-vi;PDBTitle: crystal structure of myosin vi in complex with dab2 peptide
38 d1qmha1
not modelled
8.4
13
Fold: Thioredoxin foldSuperfamily: RNA 3'-terminal phosphate cyclase, RPTC, insert domainFamily: RNA 3'-terminal phosphate cyclase, RPTC, insert domain39 c2c5iP_
not modelled
8.2
21
PDB header: protein transportChain: P: PDB Molecule: vacuolar protein sorting protein 51;PDBTitle: n-terminal domain of tlg1 complexed with n-terminus of2 vps51 in distorted conformation
40 c2bcmB_
not modelled
7.9
27
PDB header: cell adhesionChain: B: PDB Molecule: f1845 fimbrial protein;PDBTitle: daae adhesin
41 d1e0ga_
not modelled
7.8
22
Fold: LysM domainSuperfamily: LysM domainFamily: LysM domain42 d1z67a1
not modelled
7.7
18
Fold: YidB-likeSuperfamily: YidB-likeFamily: YidB-like43 d1jhga_
not modelled
7.5
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: TrpR-likeFamily: Trp repressor, TrpR44 d1pn0a2
not modelled
7.5
17
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like45 c1or7A_
not modelled
7.4
14
PDB header: transcriptionChain: A: PDB Molecule: rna polymerase sigma-e factor;PDBTitle: crystal structure of escherichia coli sigmae with the cytoplasmic2 domain of its anti-sigma rsea
46 c2ia0A_
not modelled
7.3
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative hth-type transcriptional regulator pf0864;PDBTitle: transcriptional regulatory protein pf0864 from pyrococcus furiosus a2 member of the asnc family (pf0864)
47 d1ut1a_
not modelled
7.1
27
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Dr-family adhesin48 d1i1ga1
not modelled
7.1
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain49 d1h54a2
not modelled
7.1
25
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Glycosyltransferase family 36 N-terminal domain50 c1i1gA_
not modelled
7.0
11
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator lrpa;PDBTitle: crystal structure of the lrp-like transcriptional regulator from the2 archaeon pyrococcus furiosus
51 d2joka1
not modelled
6.8
40
Fold: SopE-like GEF domainSuperfamily: SopE-like GEF domainFamily: SopE-like GEF domain52 c2djpA_
not modelled
6.8
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein sb145;PDBTitle: the solution structure of the lysm domain of human2 hypothetical protein sb145
53 d1mgta1
not modelled
6.6
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domainFamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domain54 d1ofcx1
not modelled
6.5
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Myb/SANT domain55 d2a7wa1
not modelled
6.4
13
Fold: all-alpha NTP pyrophosphatasesSuperfamily: all-alpha NTP pyrophosphatasesFamily: HisE-like (PRA-PH)56 c2a7wF_
not modelled
6.4
13
PDB header: hydrolaseChain: F: PDB Molecule: phosphoribosyl-atp pyrophosphatase;PDBTitle: crystal structure of phosphoribosyl-atp pyrophosphatase2 from chromobacterium violaceum (atcc 12472). nesg target3 cvr7
57 c2opkC_
not modelled
6.1
27
PDB header: isomeraseChain: C: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a putative mannose-6-phosphate isomerase2 (reut_a1446) from ralstonia eutropha jmp134 at 2.10 a resolution
58 d1s6la1
not modelled
5.8
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MerB N-terminal domain-like59 d2vera1
not modelled
5.8
20
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Dr-family adhesin60 c2vbzA_
not modelled
5.5
23
PDB header: dna-binding proteinChain: A: PDB Molecule: transcriptional regulatory protein;PDBTitle: feast or famine regulatory protein (rv3291c)from m.2 tuberculosis complexed with l-tryptophan
61 c2krcA_
not modelled
5.4
12
PDB header: transcriptionChain: A: PDB Molecule: dna-directed rna polymerase subunit delta;PDBTitle: solution structure of the n-terminal domain of bacillus2 subtilis delta subunit of rna polymerase
62 c2l4aA_
not modelled
5.4
11
PDB header: dna binding proteinChain: A: PDB Molecule: leucine responsive regulatory protein;PDBTitle: nmr structure of the dna-binding domain of e.coli lrp
63 d1or7a1
not modelled
5.1
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain64 d2cfxa1
not modelled
5.1
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain