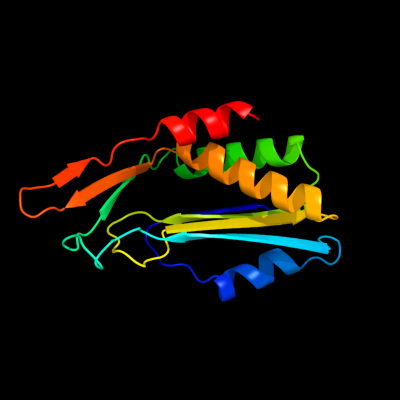
| 1 | d1fm0e_
|
|
 |
100.0 |
100 |
Fold:alpha/beta-Hammerhead
Superfamily:Molybdopterin synthase subunit MoaE
Family:Molybdopterin synthase subunit MoaE |
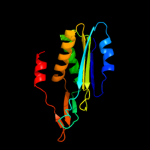
| 2 | c2qieA_
|
|
 |
100.0 |
32 |
PDB header:transferase
Chain: A: PDB Molecule:molybdopterin-converting factor subunit 2;
PDBTitle: staphylococcus aureus molybdopterin synthase in complex2 with precursor z
|
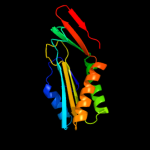
| 3 | c2omdB_
|
|
 |
100.0 |
29 |
PDB header:lyase
Chain: B: PDB Molecule:molybdopterin-converting factor subunit 2;
PDBTitle: crystal structure of molybdopterin converting factor subunit 22 (aq_2181) from aquifex aeolicus vf5
|
| 4 | c3rpfB_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:molybdopterin synthase catalytic subunit;
PDBTitle: protein-protein complex of subunit 1 and 2 of molybdopterin-converting2 factor from helicobacter pylori 26695
|
| 5 | d1nvja_
|
|
 |
100.0 |
100 |
Fold:alpha/beta-Hammerhead
Superfamily:Molybdopterin synthase subunit MoaE
Family:Molybdopterin synthase subunit MoaE |
| 6 | c2wp4A_
|
|
 |
100.0 |
36 |
PDB header:transferase
Chain: A: PDB Molecule:molybdopterin-converting factor subunit 2 1;
PDBTitle: crystal structure of rv3119 from mycobacterium tuberculosis
|
| 7 | c1v8cA_
|
|
 |
98.8 |
21 |
PDB header:protein binding
Chain: A: PDB Molecule:moad related protein;
PDBTitle: crystal structure of moad related protein from thermus2 thermophilus hb8
|
| 8 | c3upsA_
|
|
 |
78.0 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:iojap-like protein;
PDBTitle: crystal structure of iojap-like protein from zymomonas mobilis
|
| 9 | d2id1a1
|
|
 |
61.0 |
12 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Iojap/YbeB-like |
| 10 | d2o5aa1
|
|
 |
33.2 |
15 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Iojap/YbeB-like |
| 11 | c2ys6A_
|
|
 |
32.4 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylglycinamide synthetase;
PDBTitle: crystal structure of gar synthetase from geobacillus kaustophilus
|
| 12 | d1n0ua5
|
|
 |
27.8 |
24 |
Fold:Ferredoxin-like
Superfamily:EF-G C-terminal domain-like
Family:EF-G/eEF-2 domains III and V |
| 13 | d2dy1a5
|
|
 |
27.4 |
20 |
Fold:Ferredoxin-like
Superfamily:EF-G C-terminal domain-like
Family:EF-G/eEF-2 domains III and V |
| 14 | c1zcoA_
|
|
 |
21.1 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphoheptonate aldolase;
PDBTitle: crystal structure of pyrococcus furiosus 3-deoxy-d-arabino-2 heptulosonate 7-phosphate synthase
|
| 15 | d1sq1a_
|
|
 |
15.4 |
9 |
Fold:Chorismate synthase, AroC
Superfamily:Chorismate synthase, AroC
Family:Chorismate synthase, AroC |
| 16 | c2i2xO_
|
|
 |
13.4 |
17 |
PDB header:transferase
Chain: O: PDB Molecule:methyltransferase 1;
PDBTitle: crystal structure of methanol:cobalamin methyltransferase complex2 mtabc from methanosarcina barkeri
|
| 17 | d1xmba2
|
|
 |
13.4 |
11 |
Fold:Ferredoxin-like
Superfamily:Bacterial exopeptidase dimerisation domain
Family:Bacterial exopeptidase dimerisation domain |
| 18 | c1zrsB_
|
|
 |
13.1 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:hypothetical protein;
PDBTitle: wild-type ld-carboxypeptidase
|
| 19 | c3lvmB_
|
|
 |
13.1 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:cysteine desulfurase;
PDBTitle: crystal structure of e.coli iscs
|
| 20 | c3cb4D_
|
|
 |
12.6 |
16 |
PDB header:translation
Chain: D: PDB Molecule:gtp-binding protein lepa;
PDBTitle: the crystal structure of lepa
|
| 21 | c2kouA_ |
|
not modelled |
12.1 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:dicer-like protein 4;
PDBTitle: dicer like protein
|
| 22 | d2od6a1 |
|
not modelled |
10.6 |
26 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:Marine metagenome family DABB1 |
| 23 | d2k49a2 |
|
not modelled |
10.5 |
8 |
Fold:YegP-like
Superfamily:YegP-like
Family:YegP-like |
| 24 | d2dkya1 |
|
not modelled |
9.9 |
33 |
Fold:SAM domain-like
Superfamily:SAM/Pointed domain
Family:Variant SAM domain |
| 25 | d1p3wa_ |
|
not modelled |
9.7 |
17 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 26 | d1uwda_ |
|
not modelled |
9.4 |
12 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Fe-S cluster assembly (FSCA) domain-like
Family:PaaD-like |
| 27 | d1rpma_ |
|
not modelled |
8.6 |
17 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Higher-molecular-weight phosphotyrosine protein phosphatases |
| 28 | c2xd4A_ |
|
not modelled |
8.5 |
13 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylamine--glycine ligase;
PDBTitle: nucleotide-bound structures of bacillus subtilis glycinamide2 ribonucleotide synthetase
|
| 29 | c3gjzB_ |
|
not modelled |
8.0 |
21 |
PDB header:immune system
Chain: B: PDB Molecule:microcin immunity protein mccf;
PDBTitle: crystal structure of microcin immunity protein mccf from bacillus2 anthracis str. ames
|
| 30 | d2qn6b1 |
|
not modelled |
7.8 |
26 |
Fold:Ferredoxin-like
Superfamily:eIF-2-alpha, C-terminal domain
Family:eIF-2-alpha, C-terminal domain |
| 31 | d2k8ea1 |
|
not modelled |
7.6 |
7 |
Fold:YegP-like
Superfamily:YegP-like
Family:YegP-like |
| 32 | d2burb1 |
|
not modelled |
7.4 |
24 |
Fold:Prealbumin-like
Superfamily:Aromatic compound dioxygenase
Family:Aromatic compound dioxygenase |
| 33 | d2k49a1 |
|
not modelled |
7.3 |
7 |
Fold:YegP-like
Superfamily:YegP-like
Family:YegP-like |
| 34 | d1dt9a3 |
|
not modelled |
7.3 |
10 |
Fold:N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1
Superfamily:N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1
Family:N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1 |
| 35 | c3iuuA_ |
|
not modelled |
7.2 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative metallopeptidase;
PDBTitle: crystal structure of putative metallopeptidase (yp_676511.1) from2 mesorhizobium sp. bnc1 at 2.13 a resolution
|
| 36 | d1larb1 |
|
not modelled |
7.2 |
14 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Higher-molecular-weight phosphotyrosine protein phosphatases |
| 37 | d2k8ea2 |
|
not modelled |
7.2 |
7 |
Fold:YegP-like
Superfamily:YegP-like
Family:YegP-like |
| 38 | d2h80a1 |
|
not modelled |
7.1 |
21 |
Fold:SAM domain-like
Superfamily:SAM/Pointed domain
Family:Variant SAM domain |
| 39 | c2bijA_ |
|
not modelled |
6.9 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:tyrosine-protein phosphatase, non-receptor type 5;
PDBTitle: crystal structure of the human protein tyrosine phosphatase2 ptpn5 (step, striatum enriched enriched phosphatase)
|
| 40 | c3okqA_ |
|
not modelled |
6.9 |
22 |
PDB header:protein binding
Chain: A: PDB Molecule:bud site selection protein 6;
PDBTitle: crystal structure of a core domain of yeast actin nucleation cofactor2 bud6
|
| 41 | d1udsa2 |
|
not modelled |
6.8 |
17 |
Fold:Ribonuclease PH domain 2-like
Superfamily:Ribonuclease PH domain 2-like
Family:Ribonuclease PH domain 2-like |
| 42 | d3bida1 |
|
not modelled |
6.8 |
27 |
Fold:YegP-like
Superfamily:YegP-like
Family:YegP-like |
| 43 | d2auna2 |
|
not modelled |
6.8 |
14 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:LD-carboxypeptidase A N-terminal domain-like |
| 44 | d2pa2a1 |
|
not modelled |
6.7 |
15 |
Fold:alpha/beta-Hammerhead
Superfamily:Ribosomal protein L16p/L10e
Family:Ribosomal protein L10e |
| 45 | c3eggC_ |
|
not modelled |
6.6 |
40 |
PDB header:hydrolase
Chain: C: PDB Molecule:spinophilin;
PDBTitle: crystal structure of a complex between protein phosphatase 1 alpha2 (pp1) and the pp1 binding and pdz domains of spinophilin
|
| 46 | c2h04A_ |
|
not modelled |
6.5 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein tyrosine phosphatase, receptor type, b,;
PDBTitle: structural studies of protein tyrosine phosphatase beta2 catalytic domain in complex with inhibitors
|
| 47 | c3e15D_ |
|
not modelled |
6.4 |
12 |
PDB header:hydrolase
Chain: D: PDB Molecule:glucose-6-phosphate 1-dehydrogenase;
PDBTitle: 6-phosphogluconolactonase from plasmodium vivax
|
| 48 | c1dd3D_ |
|
not modelled |
6.1 |
24 |
PDB header:ribosome
Chain: D: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: crystal structure of ribosomal protein l12 from thermotoga maritima
|
| 49 | c1dd3C_ |
|
not modelled |
6.1 |
24 |
PDB header:ribosome
Chain: C: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: crystal structure of ribosomal protein l12 from thermotoga maritima
|
| 50 | d1v6ba_ |
|
not modelled |
6.1 |
40 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 51 | d1p15a_ |
|
not modelled |
6.1 |
18 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Higher-molecular-weight phosphotyrosine protein phosphatases |
| 52 | c2i75A_ |
|
not modelled |
6.1 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:tyrosine-protein phosphatase non-receptor type 4;
PDBTitle: crystal structure of human protein tyrosine phosphatase n4 (ptpn4)
|
| 53 | d2pjua1 |
|
not modelled |
5.9 |
16 |
Fold:Chelatase-like
Superfamily:PrpR receptor domain-like
Family:PrpR receptor domain-like |
| 54 | d2o0ma1 |
|
not modelled |
5.9 |
21 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
| 55 | c2o0mA_ |
|
not modelled |
5.9 |
21 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, sorc family;
PDBTitle: the crystal structure of the putative sorc family transcriptional2 regulator from enterococcus faecalis
|
| 56 | c1ceuA_ |
|
not modelled |
5.8 |
35 |
PDB header:viral protein
Chain: A: PDB Molecule:protein (hiv-1 regulatory protein n-terminal
PDBTitle: nmr structure of the (1-51) n-terminal domain of the hiv-12 regulatory protein
|
| 57 | c2pjuD_ |
|
not modelled |
5.8 |
16 |
PDB header:transcription
Chain: D: PDB Molecule:propionate catabolism operon regulatory protein;
PDBTitle: crystal structure of propionate catabolism operon2 regulatory protein prpr
|
| 58 | d2gnoa1 |
|
not modelled |
5.7 |
6 |
Fold:post-AAA+ oligomerization domain-like
Superfamily:post-AAA+ oligomerization domain-like
Family:DNA polymerase III clamp loader subunits, C-terminal domain |
| 59 | d1jlna_ |
|
not modelled |
5.6 |
21 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Higher-molecular-weight phosphotyrosine protein phosphatases |
| 60 | d1wfga_ |
|
not modelled |
5.4 |
40 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 61 | d1yfoa_ |
|
not modelled |
5.4 |
16 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Higher-molecular-weight phosphotyrosine protein phosphatases |
| 62 | c2kl8A_ |
|
not modelled |
5.4 |
19 |
PDB header:de novo protein
Chain: A: PDB Molecule:or15;
PDBTitle: solution nmr structure of de novo designed ferredoxin-like2 fold protein, northeast structural genomics consortium3 target or15
|
| 63 | c3jrlA_ |
|
not modelled |
5.4 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:oncogenic tyrosine phosphatase shp2;
PDBTitle: crystal structure of the oncogenic tyrosine phosphatase shp2 complexed2 with a salicylic acid-based small molecule inhibitor
|
| 64 | c2qdmA_ |
|
not modelled |
5.4 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:tyrosine-protein phosphatase non-receptor type 7;
PDBTitle: crystal structure of the heptp catalytic domain c270s/d236a/q314a2 mutant
|
| 65 | d2k7ia1 |
|
not modelled |
5.4 |
7 |
Fold:YegP-like
Superfamily:YegP-like
Family:YegP-like |
| 66 | c2k7iB_ |
|
not modelled |
5.4 |
7 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:upf0339 protein atu0232;
PDBTitle: solution nmr structure of protein atu0232 from agrobacterium2 tumefaciens. northeast structural genomics consortium (nesg) target3 att3. ontario center for structural proteomics target atc0223.
|
| 67 | c2b49A_ |
|
not modelled |
5.2 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein tyrosine phosphatase, non-receptor type 3;
PDBTitle: crystal structure of the catalytic domain of protein tyrosine2 phosphatase, non-receptor type 3
|