| 1 |
|
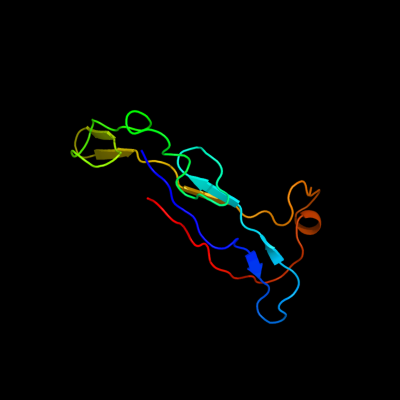
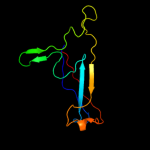
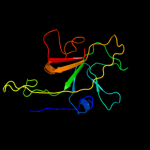
PDB 3f85 chain A
Region: 85 - 190
Aligned: 86
Modelled: 101
Confidence: 43.4%
Identity: 22%
PDB header:cell adhesion
Chain: A: PDB Molecule:homo trimeric fusion of cfa/i fimbrial subunits b;
PDBTitle: structure of fusion complex of homo trimeric major pilin subunits cfab2 of cfa/i fimbirae from etec e. coli
Phyre2
| 2 |
|
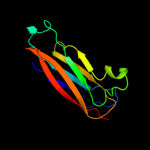
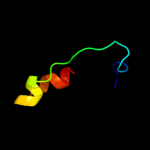
PDB 1qfh chain B
Region: 68 - 192
Aligned: 118
Modelled: 125
Confidence: 38.3%
Identity: 17%
PDB header:actin binding protein
Chain: B: PDB Molecule:protein (gelation factor);
PDBTitle: dimerization of gelation factor from dictyostelium2 discoideum: crystal structure of rod domains 5 and 6
Phyre2
| 3 |
|
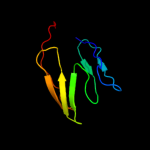
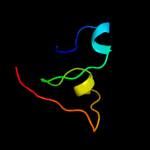
PDB 1ofs chain C
Region: 51 - 190
Aligned: 116
Modelled: 130
Confidence: 30.3%
Identity: 22%
PDB header:lectin
Chain: C: PDB Molecule:pea lectin alpha chain;
PDBTitle: pea lectin-sucrose complex
Phyre2
| 4 |
|
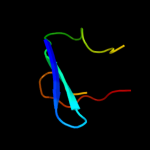
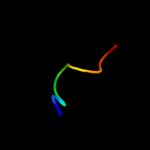
PDB 3f84 chain A
Region: 86 - 190
Aligned: 86
Modelled: 105
Confidence: 22.9%
Identity: 23%
PDB header:cell adhesion
Chain: A: PDB Molecule:cfa/i fimbrial subunit b;
PDBTitle: structure of fusion complex of major pilin cfab and major pilin cfab2 of cfa/i pilus from etec e. coli
Phyre2
| 5 |
|
PDB 3f83 chain A
Region: 35 - 190
Aligned: 132
Modelled: 143
Confidence: 20.9%
Identity: 23%
PDB header:cell adhesion
Chain: A: PDB Molecule:fusion of the minor pilin cfae and major pilin cfab;
PDBTitle: structure of fusion complex of the minor pilin cfae and major pilin2 cfab of cfa/i pili from etec e. coli
Phyre2
| 6 |
|
PDB 1cwv chain A domain 1
Region: 84 - 191
Aligned: 92
Modelled: 108
Confidence: 18.6%
Identity: 24%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: Invasin/intimin cell-adhesion fragments
Family: Invasin/intimin cell-adhesion fragments
Phyre2
| 7 |
|
PDB 2qqp chain G
Region: 49 - 127
Aligned: 78
Modelled: 79
Confidence: 12.1%
Identity: 28%
PDB header:virus
Chain: G: PDB Molecule:large capsid protein;
PDBTitle: crystal structure of authentic providence virus
Phyre2
| 8 |
|
PDB 1tg7 chain A domain 1
Region: 87 - 119
Aligned: 33
Modelled: 33
Confidence: 10.6%
Identity: 24%
Fold: Beta-galactosidase LacA, domain 3
Superfamily: Beta-galactosidase LacA, domain 3
Family: Beta-galactosidase LacA, domain 3
Phyre2
| 9 |
|
PDB 1g7y chain A
Region: 51 - 190
Aligned: 120
Modelled: 140
Confidence: 8.2%
Identity: 18%
Fold: Concanavalin A-like lectins/glucanases
Superfamily: Concanavalin A-like lectins/glucanases
Family: Legume lectins
Phyre2
| 10 |
|
PDB 2c5s chain A
Region: 111 - 141
Aligned: 26
Modelled: 31
Confidence: 7.7%
Identity: 31%
PDB header:rna-binding protein
Chain: A: PDB Molecule:probable thiamine biosynthesis protein thii;
PDBTitle: crystal structure of bacillus anthracis thii, a trna-2 modifying enzyme containing the predicted rna-binding3 thump domain
Phyre2
| 11 |
|
PDB 1q14 chain A
Region: 43 - 78
Aligned: 36
Modelled: 36
Confidence: 6.9%
Identity: 25%
PDB header:hydrolase
Chain: A: PDB Molecule:hst2 protein;
PDBTitle: structure and autoregulation of the yeast hst2 homolog of sir2
Phyre2
| 12 |
|
PDB 2jsh chain A
Region: 68 - 76
Aligned: 9
Modelled: 9
Confidence: 6.3%
Identity: 44%
PDB header:hormone
Chain: A: PDB Molecule:appetite-regulating hormone, obestatin;
PDBTitle: obestatin nmr structure in sds/dpc micellar solution
Phyre2