| 1 |
|
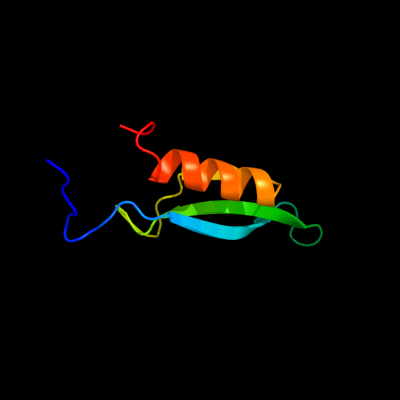
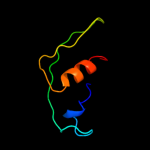
PDB 2kvt chain A
Region: 1 - 63
Aligned: 63
Modelled: 63
Confidence: 100.0%
Identity: 100%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein yaia;
PDBTitle: solution nmr structure of yaia from escherichia eoli. northeast2 structural genomics target er244
Phyre2
| 2 |
|
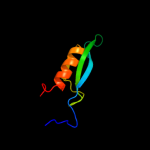
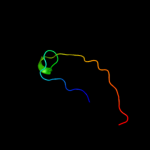
PDB 2krx chain A
Region: 21 - 36
Aligned: 16
Modelled: 16
Confidence: 27.8%
Identity: 25%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:asl3597 protein;
PDBTitle: solution nmr structure of asl3597 from nostoc sp. pcc7120.2 northeast structural genomics consortium target id nsr244.
Phyre2
| 3 |
|
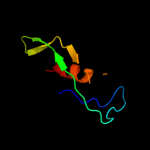
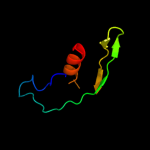
PDB 2gsk chain B domain 1
Region: 4 - 57
Aligned: 50
Modelled: 54
Confidence: 27.0%
Identity: 22%
Fold: TolA/TonB C-terminal domain
Superfamily: TolA/TonB C-terminal domain
Family: TonB
Phyre2
| 4 |
|
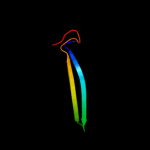
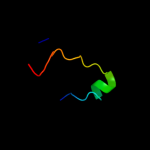
PDB 1j6w chain A
Region: 4 - 40
Aligned: 34
Modelled: 37
Confidence: 25.9%
Identity: 32%
Fold: LuxS/MPP-like metallohydrolase
Superfamily: LuxS/MPP-like metallohydrolase
Family: Autoinducer-2 production protein LuxS
Phyre2
| 5 |
|
PDB 1j98 chain A
Region: 4 - 40
Aligned: 34
Modelled: 37
Confidence: 25.8%
Identity: 18%
Fold: LuxS/MPP-like metallohydrolase
Superfamily: LuxS/MPP-like metallohydrolase
Family: Autoinducer-2 production protein LuxS
Phyre2
| 6 |
|
PDB 2grx chain C
Region: 4 - 57
Aligned: 50
Modelled: 54
Confidence: 22.6%
Identity: 22%
PDB header:metal transport
Chain: C: PDB Molecule:protein tonb;
PDBTitle: crystal structure of tonb in complex with fhua, e. coli2 outer membrane receptor for ferrichrome
Phyre2
| 7 |
|
PDB 2qqd chain G
Region: 7 - 42
Aligned: 36
Modelled: 36
Confidence: 15.3%
Identity: 31%
PDB header:lyase
Chain: G: PDB Molecule:pyruvoyl-dependent arginine decarboxylase (ec
PDBTitle: n47a mutant of pyruvoyl-dependent arginine decarboxylase2 from methanococcus jannashii
Phyre2
| 8 |
|
PDB 1u07 chain A
Region: 3 - 33
Aligned: 27
Modelled: 31
Confidence: 14.9%
Identity: 26%
Fold: TolA/TonB C-terminal domain
Superfamily: TolA/TonB C-terminal domain
Family: TonB
Phyre2
| 9 |
|
PDB 3t6o chain A
Region: 38 - 53
Aligned: 16
Modelled: 16
Confidence: 12.1%
Identity: 38%
PDB header:transport protein
Chain: A: PDB Molecule:sulfate transporter/antisigma-factor antagonist stas;
PDBTitle: the structure of an anti-sigma-factor antagonist (stas) domain protein2 from planctomyces limnophilus.
Phyre2
| 10 |
|
PDB 1xx3 chain A
Region: 4 - 57
Aligned: 50
Modelled: 54
Confidence: 11.9%
Identity: 22%
PDB header:transport protein
Chain: A: PDB Molecule:tonb protein;
PDBTitle: solution structure of escherichia coli tonb-ctd
Phyre2
| 11 |
|
PDB 1jj2 chain H
Region: 4 - 30
Aligned: 23
Modelled: 27
Confidence: 11.5%
Identity: 43%
Fold: alpha/beta-Hammerhead
Superfamily: Ribosomal protein L16p/L10e
Family: Ribosomal protein L10e
Phyre2
| 12 |
|
PDB 1n13 chain J
Region: 7 - 42
Aligned: 36
Modelled: 36
Confidence: 11.2%
Identity: 31%
PDB header:lyase
Chain: J: PDB Molecule:pyruvoyl-dependent arginine decarboxylase alpha
PDBTitle: the crystal structure of pyruvoyl-dependent arginine2 decarboxylase from methanococcus jannashii
Phyre2
| 13 |
|
PDB 4a1a chain H
Region: 4 - 30
Aligned: 23
Modelled: 27
Confidence: 9.1%
Identity: 30%
PDB header:ribosome
Chain: H: PDB Molecule:60s ribosomal protein l10;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 3.
Phyre2
| 14 |
|
PDB 1j6x chain A
Region: 12 - 40
Aligned: 29
Modelled: 28
Confidence: 6.7%
Identity: 28%
Fold: LuxS/MPP-like metallohydrolase
Superfamily: LuxS/MPP-like metallohydrolase
Family: Autoinducer-2 production protein LuxS
Phyre2
| 15 |
|
PDB 2zkd chain A domain 1
Region: 12 - 44
Aligned: 33
Modelled: 33
Confidence: 6.2%
Identity: 36%
Fold: PUA domain-like
Superfamily: PUA domain-like
Family: SRA domain-like
Phyre2
| 16 |
|
PDB 2pb7 chain A
Region: 16 - 37
Aligned: 22
Modelled: 22
Confidence: 6.0%
Identity: 50%
PDB header:ligase
Chain: A: PDB Molecule:e3 ubiquitin-protein ligase uhrf1;
PDBTitle: crystal structure of the sra domain of the human uhrf12 protein
Phyre2
| 17 |
|
PDB 3k6q chain B
Region: 40 - 53
Aligned: 14
Modelled: 14
Confidence: 5.5%
Identity: 21%
PDB header:ligand binding protein
Chain: B: PDB Molecule:putative ligand binding protein;
PDBTitle: crystal structure of an antitoxin part of a putative toxin/antitoxin2 system (swol_0700) from syntrophomonas wolfei subsp. wolfei at 1.80 a3 resolution
Phyre2
| 18 |
|
PDB 3bi7 chain A domain 1
Region: 16 - 43
Aligned: 28
Modelled: 28
Confidence: 5.3%
Identity: 43%
Fold: PUA domain-like
Superfamily: PUA domain-like
Family: SRA domain-like
Phyre2
| 19 |
|
PDB 1ffk chain F
Region: 4 - 30
Aligned: 23
Modelled: 27
Confidence: 5.0%
Identity: 43%
Fold: alpha/beta-Hammerhead
Superfamily: Ribosomal protein L16p/L10e
Family: Ribosomal protein L10e
Phyre2