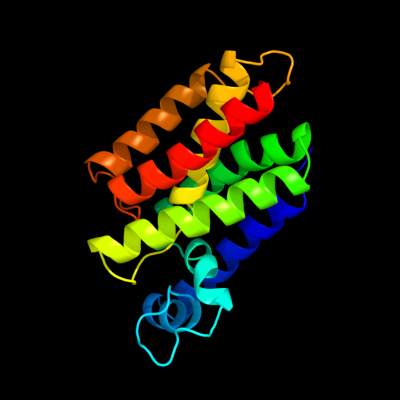
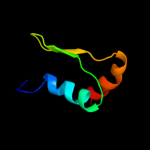
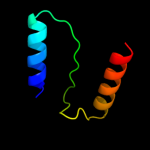
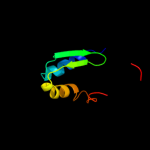
1 d3b45a1
100.0
100
Fold: Rhomboid-likeSuperfamily: Rhomboid-likeFamily: Rhomboid-like2 d2nr9a1
100.0
36
Fold: Rhomboid-likeSuperfamily: Rhomboid-likeFamily: Rhomboid-like3 c2gqcA_
98.6
25
PDB header: hydrolaseChain: A: PDB Molecule: rhomboid intramembrane protease;PDBTitle: solution structure of the n-terminal domain of rhomboid intramembrane2 protease from p. aeruginosa
4 d1utaa_
93.9
19
Fold: Ferredoxin-likeSuperfamily: Sporulation related repeatFamily: Sporulation related repeat5 c1x60A_
93.5
15
PDB header: hydrolaseChain: A: PDB Molecule: sporulation-specific n-acetylmuramoyl-l-alaninePDBTitle: solution structure of the peptidoglycan binding domain of2 b. subtilis cell wall lytic enzyme cwlc
6 c2y9jt_
93.5
4
PDB header: protein transportChain: T: PDB Molecule: protein prgh;PDBTitle: three-dimensional model of salmonella's needle complex at2 subnanometer resolution
7 c1yj7A_
92.7
9
PDB header: protein transportChain: A: PDB Molecule: escj;PDBTitle: crystal structure of enteropathogenic e.coli (epec) type iii secretion2 system protein escj
8 d1zhva2
60.8
16
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Atu0741-like9 c2hfvA_
53.8
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein rpa1041;PDBTitle: solution nmr structure of protein rpa1041 from pseudomonas2 aeruginosa. northeast structural genomics consortium3 target pat90.
10 d1zvpa2
51.5
18
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: VC0802-like11 d2hmfa2
44.3
18
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Aspartokinase allosteric domain-like12 d2hfva1
37.7
15
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: RPA1041-like13 c1zvpB_
30.6
18
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein vc0802;PDBTitle: crystal structure of a protein of unknown function vc0802 from vibrio2 cholerae, possible transport protein
14 d2bgra2
29.2
10
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: DPP6 catalytic domain-like15 c3jw8A_
28.1
12
PDB header: hydrolaseChain: A: PDB Molecule: mgll protein;PDBTitle: crystal structure of human mono-glyceride lipase
16 c3hjuB_
27.5
12
PDB header: hydrolaseChain: B: PDB Molecule: monoglyceride lipase;PDBTitle: crystal structure of human monoglyceride lipase
17 c1zhvA_
23.8
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein atu0741;PDBTitle: x-ray crystal structure protein atu0741 from agobacterium tumefaciens.2 northeast structural genomics consortium target atr8.
18 c2v1nA_
22.7
11
PDB header: nuclear proteinChain: A: PDB Molecule: protein kin homolog;PDBTitle: solution structure of the region 51-160 of human kin172 reveals a winged helix fold
19 d1orva2
22.4
12
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: DPP6 catalytic domain-like20 c3l76B_
19.8
25
PDB header: transferaseChain: B: PDB Molecule: aspartokinase;PDBTitle: crystal structure of aspartate kinase from synechocystis
21 c2wirB_
not modelled
19.2
9
PDB header: hydrolaseChain: B: PDB Molecule: alpha/beta hydrolase fold-3 domain protein;PDBTitle: hyperthermophilic esterase from the archeon pyrobaculum2 calidifontis
22 d1u4na_
not modelled
19.2
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carboxylesterase23 c2dtjA_
not modelled
18.2
16
PDB header: transferaseChain: A: PDB Molecule: aspartokinase;PDBTitle: crystal structure of regulatory subunit of aspartate kinase2 from corynebacterium glutamicum
24 c3qwuA_
not modelled
17.6
10
PDB header: ligaseChain: A: PDB Molecule: dna ligase;PDBTitle: putative atp-dependent dna ligase from aquifex aeolicus.
25 d2cdqa3
not modelled
15.8
19
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Aspartokinase allosteric domain-like26 d1sbpa_
not modelled
15.1
27
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like27 c3d7rB_
not modelled
14.9
8
PDB header: hydrolaseChain: B: PDB Molecule: esterase;PDBTitle: crystal structure of a putative esterase from staphylococcus aureus
28 d1xfda2
not modelled
14.2
6
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: DPP6 catalytic domain-like29 c1yq3C_
not modelled
12.8
10
PDB header: oxidoreductaseChain: C: PDB Molecule: succinate dehydrogenase cytochrome b, large subunit;PDBTitle: avian respiratory complex ii with oxaloacetate and ubiquinone
30 c2voyG_
not modelled
12.4
23
PDB header: hydrolaseChain: G: PDB Molecule: sarcoplasmic/endoplasmic reticulum calciumPDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
31 d2hu7a2
not modelled
12.0
8
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Acylamino-acid-releasing enzyme, C-terminal donain32 c3aikB_
not modelled
11.2
12
PDB header: hydrolaseChain: B: PDB Molecule: 303aa long hypothetical esterase;PDBTitle: crystal structure of a hsl-like carboxylesterase from sulfolobus2 tokodaii
33 d2boaa2
not modelled
11.1
22
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Pancreatic carboxypeptidase, activation domain34 c3gr0D_
not modelled
10.7
11
PDB header: membrane proteinChain: D: PDB Molecule: protein prgh;PDBTitle: periplasmic domain of the t3ss inner membrane protein prgh from2 s.typhimurium (fragment 170-362)
35 d1jqga2
not modelled
10.6
12
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Pancreatic carboxypeptidase, activation domain36 c2re1A_
not modelled
10.0
22
PDB header: transferaseChain: A: PDB Molecule: aspartokinase, alpha and beta subunits;PDBTitle: crystal structure of aspartokinase alpha and beta subunits
37 c1nauA_
not modelled
8.8
36
PDB header: hormone/growth factorChain: A: PDB Molecule: glucagon;PDBTitle: nmr solution structure of the glucagon antagonist [deshis1,2 desphe6, glu9]glucagon amide in the presence of3 perdeuterated dodecylphosphocholine micelles
38 d1tc3c_
not modelled
8.5
27
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain39 c3fakA_
not modelled
8.2
8
PDB header: hydrolaseChain: A: PDB Molecule: esterase/lipase;PDBTitle: structural and functional analysis of a hormone-sensitive2 lipase like este5 from a metagenome library
40 c2ecfA_
not modelled
8.1
10
PDB header: hydrolaseChain: A: PDB Molecule: dipeptidyl peptidase iv;PDBTitle: crystal structure of dipeptidyl aminopeptidase iv from2 stenotrophomonas maltophilia
41 c3ga7A_
not modelled
7.7
9
PDB header: hydrolaseChain: A: PDB Molecule: acetyl esterase;PDBTitle: 1.55 angstrom crystal structure of an acetyl esterase from salmonella2 typhimurium
42 d1udxa3
not modelled
7.6
20
Fold: Obg GTP-binding protein C-terminal domainSuperfamily: Obg GTP-binding protein C-terminal domainFamily: Obg GTP-binding protein C-terminal domain43 d1o6xa_
not modelled
7.5
21
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Pancreatic carboxypeptidase, activation domain44 c2k9yB_
not modelled
7.2
40
PDB header: transferaseChain: B: PDB Molecule: ephrin type-a receptor 2;PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
45 c2k9yA_
not modelled
7.2
40
PDB header: transferaseChain: A: PDB Molecule: ephrin type-a receptor 2;PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
46 c1bh0A_
not modelled
6.4
33
PDB header: synthetic hormoneChain: A: PDB Molecule: glucagon;PDBTitle: structure of a glucagon analog
47 c1jrjA_
not modelled
6.2
7
PDB header: hormone/growth factorChain: A: PDB Molecule: exendin-4;PDBTitle: solution structure of exendin-4 in 30-vol% trifluoroethanol
48 d1h97a_
not modelled
6.2
5
Fold: Globin-likeSuperfamily: Globin-likeFamily: Globins49 d2hmfa3
not modelled
5.8
10
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Aspartokinase allosteric domain-like50 d2c0ra1
not modelled
5.6
12
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like51 d1lzla_
not modelled
5.6
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carboxylesterase52 c1d0rA_
not modelled
5.4
17
PDB header: hormone/growth factorChain: A: PDB Molecule: glucagon-like peptide-1-(7-36)-amide;PDBTitle: solution structure of glucagon-like peptide-1-(7-36)-amide2 in trifluoroethanol/water
53 c2zhoB_
not modelled
5.3
13
PDB header: transferaseChain: B: PDB Molecule: aspartokinase;PDBTitle: crystal structure of the regulatory subunit of aspartate2 kinase from thermus thermophilus (ligand free form)
54 c3ofeB_
not modelled
5.3
18
PDB header: chaperoneChain: B: PDB Molecule: ldlr chaperone boca;PDBTitle: structured domain of drosophila melanogaster boca p41 2 2 crystal form