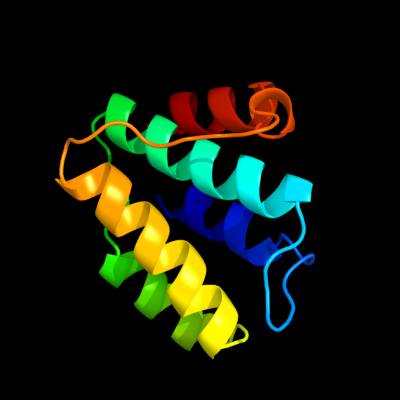
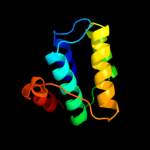
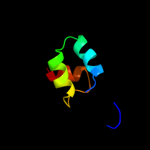
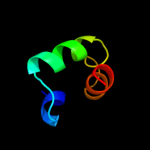
1 d2es9a1
100.0
63
Fold: YoaC-likeSuperfamily: YoaC-likeFamily: YoaC-like2 c2cosA_
55.2
35
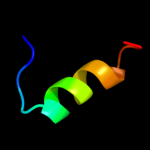
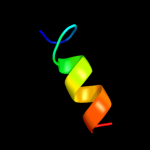
PDB header: transferaseChain: A: PDB Molecule: serine/threonine protein kinase lats2;PDBTitle: solution structure of rsgi ruh-038, a uba domain from mouse2 lats2 (large tumor suppressor homolog 2)
3 d2cosa1
43.5
45
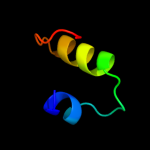
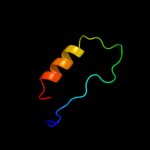
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain4 d1t8sa_
33.0
35
Fold: Phosphorylase/hydrolase-likeSuperfamily: Purine and uridine phosphorylasesFamily: Purine and uridine phosphorylases5 c2q62A_
25.3
16
PDB header: flavoproteinChain: A: PDB Molecule: arsh;PDBTitle: crystal structure of arsh from sinorhizobium meliloti
6 d1dpta_
23.4
20
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: MIF-related7 d2v4ja3
17.8
12
Fold: Nitrite and sulphite reductase 4Fe-4S domain-likeSuperfamily: Nitrite and sulphite reductase 4Fe-4S domain-likeFamily: Nitrite and sulphite reductase 4Fe-4S domain-like8 d1tnsa_
16.5
32
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: Excisionase-like9 c2o03A_
14.8
29
PDB header: gene regulationChain: A: PDB Molecule: probable zinc uptake regulation protein furb;PDBTitle: crystal structure of furb from m. tuberculosis- a zinc uptake2 regulator
10 c3n70F_
14.7
26
PDB header: transport proteinChain: F: PDB Molecule: transport activator;PDBTitle: the crystal structure of the p-loop ntpase domain of the sigma-542 transport activator from e. coli to 2.8a
11 d1qpma_
14.2
44
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: Excisionase-like12 d1mzba_
14.1
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: FUR-like13 d1jbob_
10.4
25
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins14 d1opaa_
10.1
33
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like15 d1p6pa_
9.4
33
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like16 c2zv3E_
9.4
28
PDB header: hydrolaseChain: E: PDB Molecule: peptidyl-trna hydrolase;PDBTitle: crystal structure of project mj0051 from methanocaldococcus2 jannaschii dsm 2661
17 c2ph5A_
9.2
36
PDB header: transferaseChain: A: PDB Molecule: homospermidine synthase;PDBTitle: crystal structure of the homospermidine synthase hss from legionella2 pneumophila in complex with nad, northeast structural genomics target3 lgr54
18 c3em0A_
8.9
33
PDB header: lipid binding proteinChain: A: PDB Molecule: ileal bile acid-binding protein;PDBTitle: crystal structure of zebrafish ileal bile acid-bindin protein2 complexed with cholic acid (crystal form b).
19 c3civA_
8.8
21
PDB header: hydrolaseChain: A: PDB Molecule: endo-beta-1,4-mannanase;PDBTitle: crystal structure of the endo-beta-1,4-mannanase from2 alicyclobacillus acidocaldarius
20 d2ftba1
8.7
33
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like21 d1d4aa_
not modelled
8.7
30
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Quinone reductase22 d1kqwa_
not modelled
8.6
22
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like23 c3ca8B_
not modelled
8.6
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: protein ydcf;PDBTitle: crystal structure of escherichia coli ydcf, an s-adenosyl-l-methionine2 utilizing enzyme
24 d1qrda_
not modelled
8.6
27
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Quinone reductase25 d2fs6a1
not modelled
8.4
56
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like26 c4a5qC_
not modelled
8.4
20
PDB header: hydrolaseChain: C: PDB Molecule: chi1;PDBTitle: crystal structure of the chitinase chi1 fitted into the 3d structure2 of the yersinia entomophaga toxin complex
27 c3oa5A_
not modelled
8.4
20
PDB header: hydrolaseChain: A: PDB Molecule: chi1;PDBTitle: the structure of chi1, a chitinase from yersinia entomophaga
28 d1ftpa_
not modelled
8.2
44
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like29 d1rlka_
not modelled
8.2
17
Fold: Peptidyl-tRNA hydrolase IISuperfamily: Peptidyl-tRNA hydrolase IIFamily: Peptidyl-tRNA hydrolase II30 d2cbra_
not modelled
8.1
56
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like31 c3nnkC_
not modelled
8.0
10
PDB header: transferaseChain: C: PDB Molecule: ureidoglycine-glyoxylate aminotransferase;PDBTitle: biochemical and structural characterization of a ureidoglycine2 aminotransferase in the klebsiella pneumoniae uric acid catabolic3 pathway
32 d1ggla_
not modelled
8.0
33
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like33 d1tw4a_
not modelled
7.9
44
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like34 d1vyfa_
not modelled
7.7
33
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like35 c2vmlD_
not modelled
7.6
20
PDB header: photosynthesisChain: D: PDB Molecule: phycocyanin beta chain;PDBTitle: the monoclinic structure of phycocyanin from gloeobacter2 violaceus
36 d2fhfa5
not modelled
7.6
36
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain37 c2lbaA_
not modelled
7.6
33
PDB header: lipid binding proteinChain: A: PDB Molecule: babp protein;PDBTitle: solution structure of chicken ileal babp in complex with2 glycochenodeoxycholic acid
38 c2c4bB_
not modelled
7.6
21
PDB header: fusion proteinChain: B: PDB Molecule: barnase mcoeeti fusion;PDBTitle: inhibitor cystine knot protein mcoeeti fused to the2 catalytically inactive barnase mutant h102a
39 d2ogqa1
not modelled
7.6
55
Fold: Polo-box domainSuperfamily: Polo-box domainFamily: Polo-box duplicated region40 d1fdqa_
not modelled
7.6
56
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like41 d1mdca_
not modelled
7.5
33
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like42 d1g7na_
not modelled
7.4
56
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like43 d1hmsa_
not modelled
7.4
56
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like44 d1o1va_
not modelled
7.2
33
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like45 d1lpja_
not modelled
7.2
33
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like46 d1lfoa_
not modelled
7.1
33
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like47 d2f73a1
not modelled
7.1
33
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like48 c3efeC_
not modelled
7.0
50
PDB header: chaperoneChain: C: PDB Molecule: thij/pfpi family protein;PDBTitle: the crystal structure of the thij/pfpi family protein from bacillus2 anthracis
49 d1kzwa_
not modelled
7.0
33
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like50 d1pmpa_
not modelled
6.9
56
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like51 c3pptA_
not modelled
6.8
44
PDB header: lipid binding proteinChain: A: PDB Molecule: sodium-calcium exchanger;PDBTitle: rep1-nxsq fatty acid transporter
52 d1hfoa_
not modelled
6.8
32
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: MIF-related53 d1yiva1
not modelled
6.8
56
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like54 c3mwmA_
not modelled
6.7
23
PDB header: transcriptionChain: A: PDB Molecule: putative metal uptake regulation protein;PDBTitle: graded expression of zinc-responsive genes through two regulatory2 zinc-binding sites in zur
55 d1cpcb_
not modelled
6.7
25
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins56 d1ifca_
not modelled
6.6
22
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like57 c2vqpA_
not modelled
6.6
75
PDB header: viral proteinChain: A: PDB Molecule: matrix protein;PDBTitle: structure of the matrix protein from human respiratory2 syncytial virus
58 d1bwya_
not modelled
6.5
56
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like59 d1eala_
not modelled
6.5
33
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like60 d1eyxa_
not modelled
6.5
21
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins61 d2a0aa1
not modelled
6.4
44
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like62 d1o8va_
not modelled
6.4
44
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like63 d1liaa_
not modelled
6.4
18
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins64 d2hnxa1
not modelled
6.3
56
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like65 d1crba_
not modelled
6.3
44
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like66 d1b56a_
not modelled
6.3
67
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like67 d1uyra2
not modelled
6.3
43
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Biotin dependent carboxylase carboxyltransferase domain68 c2crvA_
not modelled
6.3
80
PDB header: translationChain: A: PDB Molecule: translation initiation factor if-2;PDBTitle: solution structure of c-terminal domain of mitochondrial2 translational initiationfactor 2
69 c2vofA_
not modelled
6.3
42
PDB header: apoptosisChain: A: PDB Molecule: bcl-2-related protein a1;PDBTitle: structure of mouse a1 bound to the puma bh3-domain
70 d2gdga1
not modelled
6.1
17
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: MIF-related71 d1t0ia_
not modelled
6.1
18
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: NADPH-dependent FMN reductase72 d2j2za2
not modelled
6.1
25
Fold: C2 domain-likeSuperfamily: Periplasmic chaperone C-domainFamily: Periplasmic chaperone C-domain73 c2gqsA_
not modelled
6.0
24
PDB header: ligaseChain: A: PDB Molecule: phosphoribosylaminoimidazole-succinocarboxamidePDBTitle: saicar synthetase complexed with cair-mg2+ and adp
74 c3kreA_
not modelled
6.0
15
PDB header: ligaseChain: A: PDB Molecule: phosphoribosylaminoimidazole-succinocarboxamide synthase;PDBTitle: crystal structure of phosphoribosylaminoimidazole-succinocarboxamide2 synthase from ehrlichia chaffeensis at 1.8a resolution
75 c2o8kA_
not modelled
5.8
55
PDB header: transcription/dnaChain: A: PDB Molecule: rna polymerase sigma factor rpon;PDBTitle: nmr structure of the sigma-54 rpon domain bound to the-242 promoter element
76 c2q9sA_
not modelled
5.8
56
PDB header: lipid binding proteinChain: A: PDB Molecule: fatty acid-binding protein;PDBTitle: linoleic acid bound to fatty acid binding protein 4
77 c3fwtA_
not modelled
5.8
29
PDB header: cytokineChain: A: PDB Molecule: macrophage migration inhibitory factor-likePDBTitle: crystal structure of leishmania major mif2
78 c2z02A_
not modelled
5.7
24
PDB header: ligaseChain: A: PDB Molecule: phosphoribosylaminoimidazole-succinocarboxamidePDBTitle: crystal structure of2 phosphoribosylaminoimidazolesuccinocarboxamide synthase3 wit atp from methanocaldococcus jannaschii
79 d2fzva1
not modelled
5.7
14
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: NADPH-dependent FMN reductase80 c2xtdB_
not modelled
5.5
43
PDB header: transcriptionChain: B: PDB Molecule: tbl1 f-box-like/wd repeat-containing protein tbl1x;PDBTitle: structure of the tbl1 tetramerisation domain
81 c3nraA_
not modelled
5.5
20
PDB header: transferaseChain: A: PDB Molecule: aspartate aminotransferase;PDBTitle: crystal structure of an aspartate aminotransferase (yp_354942.1) from2 rhodobacter sphaeroides 2.4.1 at 2.15 a resolution
82 d1gd0a_
not modelled
5.4
21
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: MIF-related83 c2kuaA_
not modelled
5.4
36
PDB header: apoptosisChain: A: PDB Molecule: bcl-2-like protein 10;PDBTitle: solution structure of a divergent bcl-2 protein
84 c2xteH_
not modelled
5.4
43
PDB header: transcriptionChain: H: PDB Molecule: f-box-like/wd repeat-containing protein tbl1x;PDBTitle: structure of the tbl1 tetramerisation domain
85 d1o82a_
not modelled
5.3
50
Fold: Saposin-likeSuperfamily: Bacteriocin AS-48Family: Bacteriocin AS-4886 c2fu4B_
not modelled
5.1
29
PDB header: dna binding proteinChain: B: PDB Molecule: ferric uptake regulation protein;PDBTitle: crystal structure of the dna binding domain of e.coli fur (ferric2 uptake regulator)
87 c2xa6B_
not modelled
5.0
38
PDB header: transcriptionChain: B: PDB Molecule: kh domain-containing\,rna-binding\,signalPDBTitle: structural basis for homodimerization of the src-associated2 during mitosis, 68 kd protein (sam68) qua1 domain