| 1 |
|
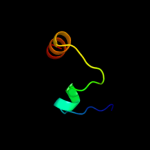
PDB 2oso chain A domain 1
Region: 55 - 86
Aligned: 32
Modelled: 29
Confidence: 49.7%
Identity: 13%
Fold: Ligand-binding domain in the NO signalling and Golgi transport
Superfamily: Ligand-binding domain in the NO signalling and Golgi transport
Family: MJ1460-like
Phyre2
| 2 |
|
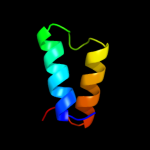
PDB 1w6k chain A domain 2
Region: 50 - 88
Aligned: 39
Modelled: 39
Confidence: 48.9%
Identity: 15%
Fold: alpha/alpha toroid
Superfamily: Terpenoid cyclases/Protein prenyltransferases
Family: Terpene synthases
Phyre2
| 3 |
|
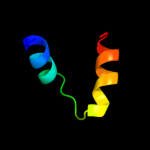
PDB 3njc chain A
Region: 55 - 86
Aligned: 32
Modelled: 28
Confidence: 30.8%
Identity: 9%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:yslb protein;
PDBTitle: crystal structure of the yslb protein from bacillus subtilis.2 northeast structural genomics consortium target sr460.
Phyre2
| 4 |
|
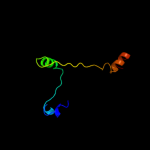
PDB 1p58 chain F
Region: 72 - 94
Aligned: 23
Modelled: 23
Confidence: 23.3%
Identity: 13%
PDB header:virus
Chain: F: PDB Molecule:envelope protein m;
PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
Phyre2
| 5 |
|
PDB 2hnh chain A
Region: 50 - 114
Aligned: 65
Modelled: 65
Confidence: 12.2%
Identity: 14%
PDB header:transferase
Chain: A: PDB Molecule:dna polymerase iii alpha subunit;
PDBTitle: crystal structure of the catalytic alpha subunit of e. coli2 replicative dna polymerase iii
Phyre2
| 6 |
|
PDB 3e0d chain A
Region: 50 - 114
Aligned: 65
Modelled: 65
Confidence: 11.5%
Identity: 23%
PDB header:transferase/dna
Chain: A: PDB Molecule:dna polymerase iii subunit alpha;
PDBTitle: insights into the replisome from the crystral structure of2 the ternary complex of the eubacterial dna polymerase iii3 alpha-subunit
Phyre2
| 7 |
|
PDB 1w6k chain A
Region: 50 - 88
Aligned: 39
Modelled: 39
Confidence: 11.2%
Identity: 15%
PDB header:isomerase
Chain: A: PDB Molecule:lanosterol synthase;
PDBTitle: structure of human osc in complex with lanosterol
Phyre2
| 8 |
|
PDB 2csb chain A domain 4
Region: 75 - 88
Aligned: 14
Modelled: 14
Confidence: 10.6%
Identity: 43%
Fold: SAM domain-like
Superfamily: RuvA domain 2-like
Family: Topoisomerase V repeat domain
Phyre2
| 9 |
|
PDB 2lbg chain A
Region: 46 - 67
Aligned: 22
Modelled: 22
Confidence: 8.3%
Identity: 32%
PDB header:membrane protein
Chain: A: PDB Molecule:major prion protein;
PDBTitle: structure of the chr of the prion protein in dpc micelles
Phyre2