| 1 |
|
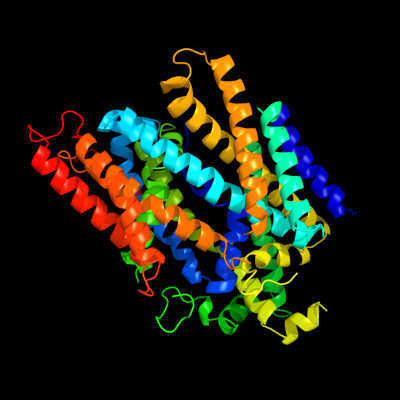
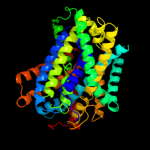
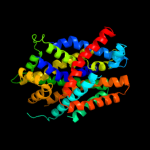
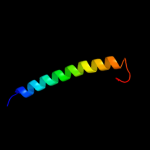
PDB 2xq2 chain A
Region: 33 - 549
Aligned: 485
Modelled: 485
Confidence: 100.0%
Identity: 16%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
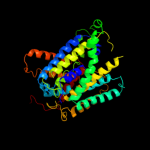
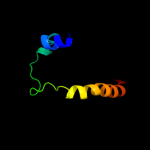
| 2 |
|
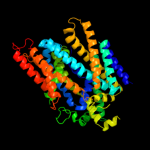
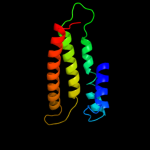
PDB 3dh4 chain A
Region: 54 - 525
Aligned: 460
Modelled: 448
Confidence: 100.0%
Identity: 14%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
Phyre2
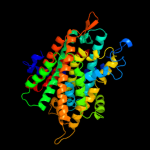
| 3 |
|
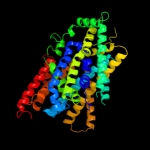
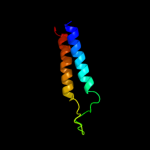
PDB 2jln chain A
Region: 66 - 540
Aligned: 431
Modelled: 431
Confidence: 99.4%
Identity: 12%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
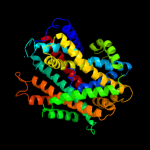
| 4 |
|
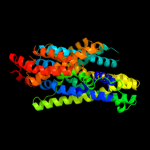
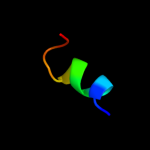
PDB 3gia chain A
Region: 67 - 526
Aligned: 423
Modelled: 423
Confidence: 99.3%
Identity: 9%
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
Phyre2
| 5 |
|
PDB 3lrc chain C
Region: 68 - 522
Aligned: 400
Modelled: 400
Confidence: 98.7%
Identity: 12%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 6 |
|
PDB 2w8a chain C
Region: 4 - 452
Aligned: 423
Modelled: 447
Confidence: 97.0%
Identity: 13%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 7 |
|
PDB 3hfx chain A
Region: 32 - 452
Aligned: 408
Modelled: 408
Confidence: 96.8%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:l-carnitine/gamma-butyrobetaine antiporter;
PDBTitle: crystal structure of carnitine transporter
Phyre2
| 8 |
|
PDB 2a65 chain A domain 1
Region: 71 - 455
Aligned: 370
Modelled: 370
Confidence: 91.0%
Identity: 14%
Fold: SNF-like
Superfamily: SNF-like
Family: SNF-like
Phyre2
| 9 |
|
PDB 3rlb chain A
Region: 410 - 523
Aligned: 112
Modelled: 114
Confidence: 88.7%
Identity: 17%
PDB header:thiamine-binding protein
Chain: A: PDB Molecule:thit;
PDBTitle: crystal structure at 2.0 a of the s-component for thiamin from an ecf-2 type abc transporter
Phyre2
| 10 |
|
PDB 3mk7 chain F
Region: 33 - 94
Aligned: 62
Modelled: 62
Confidence: 49.1%
Identity: 11%
PDB header:oxidoreductase
Chain: F: PDB Molecule:cytochrome c oxidase, cbb3-type, subunit p;
PDBTitle: the structure of cbb3 cytochrome oxidase
Phyre2
| 11 |
|
PDB 3ehb chain B domain 2
Region: 33 - 70
Aligned: 38
Modelled: 38
Confidence: 9.8%
Identity: 11%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 12 |
|
PDB 2ky5 chain A
Region: 133 - 144
Aligned: 11
Modelled: 12
Confidence: 8.0%
Identity: 18%
PDB header:cell adhesion
Chain: A: PDB Molecule:platelet endothelial cell adhesion molecule;
PDBTitle: solution structure of the pecam-1 cytoplasmic tail with dpc
Phyre2
| 13 |
|
PDB 3rko chain K
Region: 24 - 61
Aligned: 38
Modelled: 38
Confidence: 7.9%
Identity: 16%
PDB header:oxidoreductase
Chain: K: PDB Molecule:nadh-quinone oxidoreductase subunit k;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 14 |
|
PDB 2ks1 chain B
Region: 39 - 56
Aligned: 18
Modelled: 18
Confidence: 7.8%
Identity: 33%
PDB header:transferase
Chain: B: PDB Molecule:epidermal growth factor receptor;
PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
Phyre2
| 15 |
|
PDB 2k9y chain A
Region: 40 - 58
Aligned: 19
Modelled: 19
Confidence: 7.3%
Identity: 11%
PDB header:transferase
Chain: A: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
Phyre2
| 16 |
|
PDB 2k9y chain B
Region: 40 - 58
Aligned: 19
Modelled: 19
Confidence: 7.3%
Identity: 11%
PDB header:transferase
Chain: B: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
Phyre2
| 17 |
|
PDB 3dtu chain B domain 2
Region: 33 - 70
Aligned: 38
Modelled: 38
Confidence: 6.3%
Identity: 5%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 18 |
|
PDB 1si2 chain A
Region: 131 - 142
Aligned: 12
Modelled: 12
Confidence: 5.7%
Identity: 17%
Fold: SH3-like barrel
Superfamily: PAZ domain
Family: PAZ domain
Phyre2
| 19 |
|
PDB 2k21 chain A
Region: 6 - 62
Aligned: 57
Modelled: 57
Confidence: 5.6%
Identity: 14%
PDB header:membrane protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily e
PDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
Phyre2