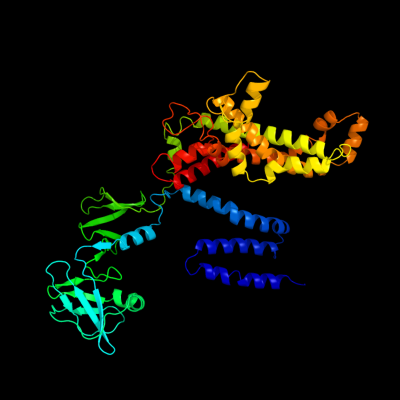
1 c2r6gF_
100.0
100
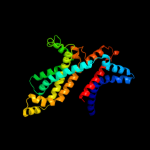
PDB header: hydrolase/transport proteinChain: F: PDB Molecule: maltose transport system permease protein malf;PDBTitle: the crystal structure of the e. coli maltose transporter
2 c3fh6F_
100.0
85
PDB header: transport proteinChain: F: PDB Molecule: maltose transport system permease protein malf;PDBTitle: crystal structure of the resting state maltose transporter from e.2 coli
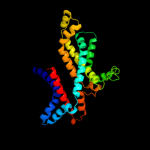
3 d2r6gf2
100.0
100
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like4 c2onkC_
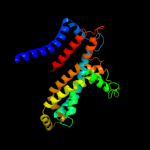
100.0
16
PDB header: membrane proteinChain: C: PDB Molecule: molybdate/tungstate abc transporter, permeasePDBTitle: abc transporter modbc in complex with its binding protein2 moda
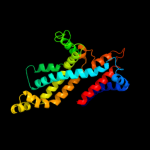
5 d2onkc1
100.0
16
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like6 c3d31D_
100.0
15
PDB header: transport proteinChain: D: PDB Molecule: sulfate/molybdate abc transporter, permeasePDBTitle: modbc from methanosarcina acetivorans
7 d3d31c1
100.0
15
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like8 d2r6gg1
100.0
18
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like9 d3dhwa1
99.8
18
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like10 d2r6gf1
99.6
100
Fold: MalF N-terminal region-likeSuperfamily: MalF N-terminal region-likeFamily: MalF N-terminal region-like11 d1ntca_
53.1
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like12 d1umqa_
50.2
3
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like13 c1umqA_
50.2
3
PDB header: dna-binding proteinChain: A: PDB Molecule: photosynthetic apparatus regulatory protein;PDBTitle: solution structure and dna binding of the effector domain2 from the global regulator prra(rega) from r. sphaeroides:3 insights into dna binding specificity
14 d1fipa_
42.4
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like15 d1etob_
37.8
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like16 d1etxa_
34.3
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like17 c3e7lD_
30.8
21
PDB header: transcription regulatorChain: D: PDB Molecule: transcriptional regulator (ntrc family);PDBTitle: crystal structure of sigma54 activator ntrc4's dna binding2 domain
18 c2hx6A_
29.0
23
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease;PDBTitle: solution structure analysis of the phage t42 endoribonuclease regb
19 d2auwa1
24.8
26
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: NE0471 C-terminal domain-like20 d1g2ha_
23.9
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like21 c2jwaA_
not modelled
15.3
15
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: erbb2 transmembrane segment dimer spatial structure
22 c2auwB_
not modelled
13.8
26
PDB header: unknown functionChain: B: PDB Molecule: hypothetical protein ne0471;PDBTitle: crystal structure of putative dna binding protein ne0471 from2 nitrosomonas europaea atcc 19718
23 d1cf7a_
not modelled
11.7
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Cell cycle transcription factor e2f-dp24 c2cw1A_
not modelled
11.5
14
PDB header: de novo proteinChain: A: PDB Molecule: sn4m;PDBTitle: solution structure of the de novo-designed lambda cro fold2 protein
25 d1wdva_
not modelled
11.4
22
Fold: YbaK/ProRS associated domainSuperfamily: YbaK/ProRS associated domainFamily: YbaK/ProRS associated domain26 c1pyuD_
not modelled
11.3
31
PDB header: lyaseChain: D: PDB Molecule: aspartate 1-decarboxylase alfa chain;PDBTitle: processed aspartate decarboxylase mutant with ser25 mutated to cys
27 d1v54g_
not modelled
11.3
18
Fold: Single transmembrane helixSuperfamily: Mitochondrial cytochrome c oxidase subunit VIaFamily: Mitochondrial cytochrome c oxidase subunit VIa28 c1iijA_
not modelled
10.9
26
PDB header: signaling proteinChain: A: PDB Molecule: erbb-2 receptor protein-tyrosine kinase;PDBTitle: solution structure of the neu/erbb-2 membrane spanning2 segment
29 c2d7dB_
not modelled
10.0
12
PDB header: hydrolase/dnaChain: B: PDB Molecule: 40-mer from uvrabc system protein b;PDBTitle: structural insights into the cryptic dna dependent atp-ase2 activity of uvrb
30 d1vz0a1
not modelled
10.0
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: KorB DNA-binding domain-likeFamily: KorB DNA-binding domain-like31 d2diia1
not modelled
9.7
22
Fold: BSD domain-likeSuperfamily: BSD domain-likeFamily: BSD domain32 d2jdih2
not modelled
9.4
11
Fold: Epsilon subunit of F1F0-ATP synthase N-terminal domainSuperfamily: Epsilon subunit of F1F0-ATP synthase N-terminal domainFamily: Epsilon subunit of F1F0-ATP synthase N-terminal domain33 c2diiA_
not modelled
9.0
22
PDB header: transcriptionChain: A: PDB Molecule: tfiih basal transcription factor complex p62PDBTitle: solution structure of the bsd domain of human tfiih basal2 transcription factor complex p62 subunit
34 c1ql1A_
not modelled
8.8
19
PDB header: virusChain: A: PDB Molecule: pf1 bacteriophage coat protein b;PDBTitle: inovirus (filamentous bacteriophage) strain pf1 major coat2 protein assembly
35 c2cx5B_
not modelled
8.7
22
PDB header: translationChain: B: PDB Molecule: a putative trans-editing enzyme;PDBTitle: crystal structure of a putative trans-editing enzyme for2 prolyl trna synthetase
36 c1vc3B_
not modelled
8.6
23
PDB header: lyaseChain: B: PDB Molecule: l-aspartate-alpha-decarboxylase heavy chain;PDBTitle: crystal structure of l-aspartate-alpha-decarboxylase
37 d2ns0a1
not modelled
8.3
7
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RHA1 ro06458-like38 c3hd7A_
not modelled
8.2
17
PDB header: exocytosisChain: A: PDB Molecule: vesicle-associated membrane protein 2;PDBTitle: helical extension of the neuronal snare complex into the membrane,2 spacegroup c 1 2 1
39 c3plxB_
not modelled
8.1
31
PDB header: lyaseChain: B: PDB Molecule: aspartate 1-decarboxylase;PDBTitle: the crystal structure of aspartate alpha-decarboxylase from2 campylobacter jejuni subsp. jejuni nctc 11168
40 c2l2tA_
not modelled
8.0
26
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-4;PDBTitle: solution nmr structure of the erbb4 dimeric membrane domain
41 c2voyG_
not modelled
7.8
18
PDB header: hydrolaseChain: G: PDB Molecule: sarcoplasmic/endoplasmic reticulum calciumPDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
42 d3fapb_
not modelled
7.5
32
Fold: Four-helical up-and-down bundleSuperfamily: FKBP12-rapamycin-binding domain of FKBP-rapamycin-associated protein (FRAP)Family: FKBP12-rapamycin-binding domain of FKBP-rapamycin-associated protein (FRAP)43 c1twcF_
not modelled
7.4
29
PDB header: transcriptionChain: F: PDB Molecule: dna-directed rna polymerases i, ii, and iii 23PDBTitle: rna polymerase ii complexed with gtp
44 d1twff_
not modelled
7.4
29
Fold: RPB6/omega subunit-likeSuperfamily: RPB6/omega subunit-likeFamily: RPB645 d1ppya_
not modelled
7.2
31
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Pyruvoyl dependent aspartate decarboxylase, ADC46 c2pmzW_
not modelled
7.2
29
PDB header: translation, transferaseChain: W: PDB Molecule: dna-directed rna polymerase subunit k;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
47 c1pt1B_
not modelled
7.2
31
PDB header: lyaseChain: B: PDB Molecule: aspartate 1-decarboxylase;PDBTitle: unprocessed pyruvoyl dependent aspartate decarboxylase with histidine2 11 mutated to alanine
48 d1st6a6
not modelled
7.1
7
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: alpha-catenin/vinculin49 d1bl0a1
not modelled
7.1
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: AraC type transcriptional activator50 c1uheA_
not modelled
7.0
23
PDB header: lyaseChain: A: PDB Molecule: aspartate 1-decarboxylase alpha chain;PDBTitle: crystal structure of aspartate decarboxylase, isoaspargine complex
51 c2k53A_
not modelled
6.9
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: a3dk08 protein;PDBTitle: nmr solution structure of a3dk08 protein from clostridium2 thermocellum: northeast structural genomics consortium3 target cmr9
52 d2axtz1
not modelled
6.9
15
Fold: Transmembrane helix hairpinSuperfamily: PsbZ-likeFamily: PsbZ-like53 c2k5eA_
not modelled
6.6
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of putative uncharacterized protein2 gsu1278 from methanocaldococcus jannaschii, northeast3 structural genomics consortium (nesg) target gsr195
54 d1jjcb2
not modelled
6.5
9
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: Domains B1 and B5 of PheRS-beta, PheT55 d1qkla_
not modelled
6.4
29
Fold: RPB6/omega subunit-likeSuperfamily: RPB6/omega subunit-likeFamily: RPB656 d1ku7a_
not modelled
6.4
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain57 d1clia1
not modelled
6.3
9
Fold: Bacillus chorismate mutase-likeSuperfamily: PurM N-terminal domain-likeFamily: PurM N-terminal domain-like58 d1clib1
not modelled
6.3
8
Fold: Bacillus chorismate mutase-likeSuperfamily: PurM N-terminal domain-likeFamily: PurM N-terminal domain-like59 d1i3ja_
not modelled
6.2
9
Fold: DNA-binding domain of intron-encoded endonucleasesSuperfamily: DNA-binding domain of intron-encoded endonucleasesFamily: DNA-binding domain of intron-encoded endonucleases60 c2i5gB_
not modelled
6.2
7
PDB header: hydrolaseChain: B: PDB Molecule: amidohydrolase;PDBTitle: crystal strcuture of amidohydrolase from pseudomonas2 aeruginosa
61 c2c45F_
not modelled
6.1
31
PDB header: lyaseChain: F: PDB Molecule: aspartate 1-decarboxylase precursor;PDBTitle: native precursor of pyruvoyl dependent aspartate2 decarboxylase
62 c3t72o_
not modelled
6.1
11
PDB header: transcription/dnaChain: O: PDB Molecule: pho box dna (strand 1);PDBTitle: phob(e)-sigma70(4)-(rnap-betha-flap-tip-helix)-dna transcription2 activation sub-complex
63 c1r71B_
not modelled
6.0
5
PDB header: transcription/dnaChain: B: PDB Molecule: transcriptional repressor protein korb;PDBTitle: crystal structure of the dna binding domain of korb in2 complex with the operator dna
64 c2g9jA_
not modelled
6.0
86
PDB header: structural proteinChain: A: PDB Molecule: tropomyosin 1 alpha chain/general control proteinPDBTitle: complex of tm1a(1-14)zip with tm9a(251-284): a model for the2 polymerization domain ("overlap region") of tropomyosin,3 northeast structural genomics target or9
65 c1l0oC_
not modelled
6.0
12
PDB header: protein bindingChain: C: PDB Molecule: sigma factor;PDBTitle: crystal structure of the bacillus stearothermophilus anti-2 sigma factor spoiiab with the sporulation sigma factor3 sigmaf
66 d1l0oc_
not modelled
6.0
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain67 c3mkuA_
not modelled
6.0
11
PDB header: transport proteinChain: A: PDB Molecule: multi antimicrobial extrusion protein (na(+)/drugPDBTitle: structure of a cation-bound multidrug and toxin compound extrusion2 (mate) transporter
68 d1coka_
not modelled
5.9
26
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain69 d1jhfa1
not modelled
5.9
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: LexA repressor, N-terminal DNA-binding domain70 c3qkbB_
not modelled
5.9
9
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a protein with unknown function which belongs to2 pfam duf74 family (pepe_0654) from pediococcus pentosaceus atcc 257453 at 2.73 a resolution
71 c3h0gF_
not modelled
5.9
29
PDB header: transcriptionChain: F: PDB Molecule: dna-directed rna polymerases i, ii, and iiiPDBTitle: rna polymerase ii from schizosaccharomyces pombe
72 d1h6gb1
not modelled
5.8
13
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: alpha-catenin/vinculin73 c3ougA_
not modelled
5.8
8
PDB header: lyaseChain: A: PDB Molecule: aspartate 1-decarboxylase;PDBTitle: crystal structure of cleaved l-aspartate-alpha-decarboxylase from2 francisella tularensis
74 c2kfsA_
not modelled
5.8
15
PDB header: dna-binding proteinChain: A: PDB Molecule: conserved hypothetical regulatory protein;PDBTitle: nmr structure of rv2175c
75 d1vr4a1
not modelled
5.7
31
Fold: Dodecin subunit-likeSuperfamily: YbjQ-likeFamily: YbjQ-like76 c3op6B_
not modelled
5.7
17
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an oligo-nucleotide binding protein (lpg1207)2 from legionella pneumophila subsp. pneumophila str. philadelphia 1 at3 2.00 a resolution
77 d1brwa3
not modelled
5.7
38
Fold: alpha/beta-HammerheadSuperfamily: Pyrimidine nucleoside phosphorylase C-terminal domainFamily: Pyrimidine nucleoside phosphorylase C-terminal domain78 c2latA_
not modelled
5.7
21
PDB header: membrane proteinChain: A: PDB Molecule: dolichyl-diphosphooligosaccharide--proteinPDBTitle: solution structure of a human minimembrane protein ost4
79 d3c9ua1
not modelled
5.6
5
Fold: Bacillus chorismate mutase-likeSuperfamily: PurM N-terminal domain-likeFamily: PurM N-terminal domain-like80 c2ka6B_
not modelled
5.6
31
PDB header: transcription regulatorChain: B: PDB Molecule: signal transducer and activator of transcriptionPDBTitle: nmr structure of the cbp-taz2/stat1-tad complex
81 d2oz6a1
not modelled
5.5
6
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like82 c1sneA_
not modelled
5.5
31
PDB header: de novo proteinChain: A: PDB Molecule: tetrameric beta-beta-alpha mini-protein;PDBTitle: an oligomeric domain-swapped beta-beta-alpha mini-protein
83 c1sneB_
not modelled
5.5
31
PDB header: de novo proteinChain: B: PDB Molecule: tetrameric beta-beta-alpha mini-protein;PDBTitle: an oligomeric domain-swapped beta-beta-alpha mini-protein
84 d1p77a2
not modelled
5.4
11
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Shikimate dehydrogenase-like85 d2tpta3
not modelled
5.4
31
Fold: alpha/beta-HammerheadSuperfamily: Pyrimidine nucleoside phosphorylase C-terminal domainFamily: Pyrimidine nucleoside phosphorylase C-terminal domain86 d2hqva1
not modelled
5.3
17
Fold: Heme iron utilization protein-likeSuperfamily: Heme iron utilization protein-likeFamily: ChuX-like87 c3rkoF_
not modelled
5.3
18
PDB header: oxidoreductaseChain: F: PDB Molecule: nadh-quinone oxidoreductase subunit j;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
88 d2phcb1
not modelled
5.3
9
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: PH0987 C-terminal domain-like89 c1y2iC_
not modelled
5.3
23
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein s0862;PDBTitle: crystal structure of mcsg target apc27401 from shigella2 flexneri
90 d1y2ia_
not modelled
5.3
23
Fold: Dodecin subunit-likeSuperfamily: YbjQ-likeFamily: YbjQ-like91 c1y6uA_
not modelled
5.3
25
PDB header: dna binding proteinChain: A: PDB Molecule: excisionase from transposon tn916;PDBTitle: the structure of the excisionase (xis) protein from2 conjugative transposon tn916 provides insights into the3 regulation of heterobivalent tyrosine recombinases
92 d1i5za1
not modelled
5.3
6
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like93 c2ph0A_
not modelled
5.3
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the q6d2t7_erwct protein from erwinia2 carotovora. nesg target ewr41.
94 c2oviA_
not modelled
5.3
6
PDB header: ligand binding protein, metal transportChain: A: PDB Molecule: hypothetical protein chux;PDBTitle: structure of the heme binding protein chux
95 d1ku2a1
not modelled
5.1
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma3 domain96 c1rkcB_
not modelled
5.1
25
PDB header: cell adhesion, structural proteinChain: B: PDB Molecule: talin;PDBTitle: human vinculin head (1-258) in complex with talin's2 vinculin binding site 3 (residues 1944-1969)
97 c2r2vB_
not modelled
5.1
38
PDB header: de novo proteinChain: B: PDB Molecule: gcn4 leucine zipper;PDBTitle: sequence determinants of the topology of the lac repressor2 tetrameric coiled coil
98 d1i1ga1
not modelled
5.0
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain99 c1jb0X_
not modelled
5.0
33
PDB header: photosynthesisChain: X: PDB Molecule: photosystem i subunit psax;PDBTitle: crystal structure of photosystem i: a photosynthetic reaction center2 and core antenna system from cyanobacteria