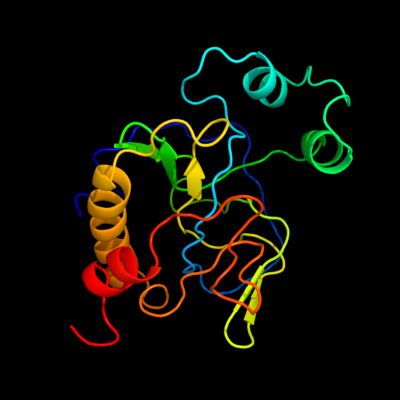
| 1 | d1j3ga_
|
|
 |
100.0 |
87 |
Fold:N-acetylmuramoyl-L-alanine amidase-like
Superfamily:N-acetylmuramoyl-L-alanine amidase-like
Family:N-acetylmuramoyl-L-alanine amidase-like |
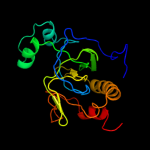
| 2 | c2bh7A_
|
|
 |
100.0 |
35 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-acetylmuramoyl-l-alanine amidase;
PDBTitle: crystal structure of a semet derivative of amid at 2.22 angstroms
|
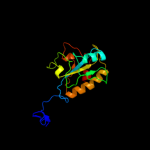
| 3 | d2bgxa2
|
|
 |
100.0 |
33 |
Fold:N-acetylmuramoyl-L-alanine amidase-like
Superfamily:N-acetylmuramoyl-L-alanine amidase-like
Family:N-acetylmuramoyl-L-alanine amidase-like |
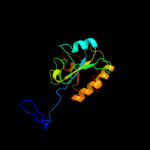
| 4 | c3hmaA_
|
|
 |
100.0 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-acetylmuramoyl-l-alanine amidase xlya;
PDBTitle: amidase from bacillus subtilis
|
| 5 | d1yb0a1
|
|
 |
100.0 |
21 |
Fold:N-acetylmuramoyl-L-alanine amidase-like
Superfamily:N-acetylmuramoyl-L-alanine amidase-like
Family:N-acetylmuramoyl-L-alanine amidase-like |
| 6 | d1ycka1
|
|
 |
100.0 |
16 |
Fold:N-acetylmuramoyl-L-alanine amidase-like
Superfamily:N-acetylmuramoyl-L-alanine amidase-like
Family:N-acetylmuramoyl-L-alanine amidase-like |
| 7 | d2cb3a1
|
|
 |
100.0 |
14 |
Fold:N-acetylmuramoyl-L-alanine amidase-like
Superfamily:N-acetylmuramoyl-L-alanine amidase-like
Family:N-acetylmuramoyl-L-alanine amidase-like |
| 8 | c2xz4A_
|
|
 |
100.0 |
16 |
PDB header:immune system
Chain: A: PDB Molecule:peptidoglycan-recognition protein lf;
PDBTitle: crystal structure of the lfz ectodomain of the2 peptidoglycan recognition protein lf
|
| 9 | c2rkqA_
|
|
 |
100.0 |
15 |
PDB header:immune system
Chain: A: PDB Molecule:peptidoglycan-recognition protein-sd;
PDBTitle: crystal structure of drosophila peptidoglycan recognition2 protein sd (pgrp-sd)
|
| 10 | d1ohta_
|
|
 |
100.0 |
17 |
Fold:N-acetylmuramoyl-L-alanine amidase-like
Superfamily:N-acetylmuramoyl-L-alanine amidase-like
Family:N-acetylmuramoyl-L-alanine amidase-like |
| 11 | c1ohtA_
|
|
 |
100.0 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:cg14704 protein;
PDBTitle: peptidoglycan recognition protein-lb
|
| 12 | d2f2lx1
|
|
 |
100.0 |
14 |
Fold:N-acetylmuramoyl-L-alanine amidase-like
Superfamily:N-acetylmuramoyl-L-alanine amidase-like
Family:N-acetylmuramoyl-L-alanine amidase-like |
| 13 | d1sk4a_
|
|
 |
100.0 |
14 |
Fold:N-acetylmuramoyl-L-alanine amidase-like
Superfamily:N-acetylmuramoyl-L-alanine amidase-like
Family:N-acetylmuramoyl-L-alanine amidase-like |
| 14 | d1lbaa_
|
|
 |
100.0 |
28 |
Fold:N-acetylmuramoyl-L-alanine amidase-like
Superfamily:N-acetylmuramoyl-L-alanine amidase-like
Family:N-acetylmuramoyl-L-alanine amidase-like |
| 15 | c3latB_
|
|
 |
100.0 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:bifunctional autolysin;
PDBTitle: crystal structure of staphylococcus peptidoglycan hydrolase2 amie
|
| 16 | c1s2jA_
|
|
 |
100.0 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidoglycan recognition protein sa cg11709-pa;
PDBTitle: crystal structure of the drosophila pattern-recognition2 receptor pgrp-sa
|
| 17 | d1sxra_
|
|
 |
100.0 |
14 |
Fold:N-acetylmuramoyl-L-alanine amidase-like
Superfamily:N-acetylmuramoyl-L-alanine amidase-like
Family:N-acetylmuramoyl-L-alanine amidase-like |
| 18 | d2f2la1
|
|
 |
100.0 |
11 |
Fold:N-acetylmuramoyl-L-alanine amidase-like
Superfamily:N-acetylmuramoyl-L-alanine amidase-like
Family:N-acetylmuramoyl-L-alanine amidase-like |
| 19 | c3ep1B_
|
|
 |
100.0 |
11 |
PDB header:immune system
Chain: B: PDB Molecule:pgrp-hd - peptidoglycan recognition protein
PDBTitle: structure of the pgrp-hd from alvinella pompejana
|
| 20 | c2xz8A_
|
|
 |
99.7 |
7 |
PDB header:immune system
Chain: A: PDB Molecule:peptidoglycan-recognition protein lf;
PDBTitle: crystal structure of the lfw ectodomain of the2 peptidoglycan recognition protein lf
|
| 21 | d2dsta1 |
|
not modelled |
50.7 |
13 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:TTHA1544-like |
| 22 | c3ds8A_ |
|
not modelled |
40.9 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lin2722 protein;
PDBTitle: the crysatl structure of the gene lin2722 products from listeria2 innocua
|
| 23 | c3pohA_ |
|
not modelled |
36.7 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:endo-beta-n-acetylglucosaminidase f1;
PDBTitle: crystal structure of an endo-beta-n-acetylglucosaminidase (bt_3987)2 from bacteroides thetaiotaomicron vpi-5482 at 1.55 a resolution
|
| 24 | c1cr6A_ |
|
not modelled |
28.1 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:epoxide hydrolase;
PDBTitle: crystal structure of murine soluble epoxide hydrolase2 complexed with cpu inhibitor
|
| 25 | c3d8mA_ |
|
not modelled |
23.7 |
36 |
PDB header:virus/viral protein
Chain: A: PDB Molecule:baseplate protein, receptor binding protein;
PDBTitle: crystal structure of a chimeric receptor binding protein from2 lactococcal phages subspecies tp901-1 and p2
|
| 26 | d1hpla2 |
|
not modelled |
23.1 |
16 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Pancreatic lipase, N-terminal domain |
| 27 | d1zrua1 |
|
not modelled |
22.1 |
36 |
Fold:Virus attachment protein globular domain
Superfamily:Virus attachment protein globular domain
Family:Lactophage receptor-binding protein head domain |
| 28 | c3da0C_ |
|
not modelled |
21.9 |
36 |
PDB header:viral protein
Chain: C: PDB Molecule:cleaved chimeric receptor binding protein from
PDBTitle: crystal structure of a cleaved form of a chimeric receptor binding2 protein from lactococcal phages subspecies tp901-1 and p2
|
| 29 | c2y96A_ |
|
not modelled |
21.4 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:dual specificity phosphatase dupd1;
PDBTitle: structure of human dual-specificity phosphatase 27
|
| 30 | c2pplA_ |
|
not modelled |
19.4 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:pancreatic lipase-related protein 1;
PDBTitle: human pancreatic lipase-related protein 1
|
| 31 | c1hplB_ |
|
not modelled |
18.6 |
16 |
PDB header:hydrolase(carboxylic esterase)
Chain: B: PDB Molecule:lipase;
PDBTitle: horse pancreatic lipase. the crystal structure at 2.32 angstroms resolution
|
| 32 | d1etha2 |
|
not modelled |
18.0 |
16 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Pancreatic lipase, N-terminal domain |
| 33 | c2pvsB_ |
|
not modelled |
17.3 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:pancreatic lipase-related protein 2;
PDBTitle: structure of human pancreatic lipase related protein 22 mutant n336q
|
| 34 | d1bu8a2 |
|
not modelled |
16.3 |
16 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Pancreatic lipase, N-terminal domain |
| 35 | d1rp1a2 |
|
not modelled |
16.1 |
24 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Pancreatic lipase, N-terminal domain |
| 36 | c1rp1A_ |
|
not modelled |
16.0 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:pancreatic lipase related protein 1;
PDBTitle: dog pancreatic lipase related protein 1
|
| 37 | c1gplA_ |
|
not modelled |
15.6 |
16 |
PDB header:serine esterase
Chain: A: PDB Molecule:rp2 lipase;
PDBTitle: rp2 lipase
|
| 38 | d1lpbb2 |
|
not modelled |
15.3 |
16 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Pancreatic lipase, N-terminal domain |
| 39 | d1xw8a_ |
|
not modelled |
13.3 |
15 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:LamB/YcsF-like |
| 40 | c3mlcC_ |
|
not modelled |
12.5 |
11 |
PDB header:isomerase
Chain: C: PDB Molecule:fg41 malonate semialdehyde decarboxylase;
PDBTitle: crystal structure of fg41msad inactivated by 3-chloropropiolate
|
| 41 | d1gpla2 |
|
not modelled |
11.4 |
13 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Pancreatic lipase, N-terminal domain |
| 42 | c2zycA_ |
|
not modelled |
10.5 |
4 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidoglycan hydrolase flgj;
PDBTitle: crystal structure of peptidoglycan hydrolase from2 sphingomonas sp. a1
|
| 43 | c3lp5A_ |
|
not modelled |
10.4 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative cell surface hydrolase;
PDBTitle: the crystal structure of the putative cell surface hydrolase from2 lactobacillus plantarum wcfs1
|
| 44 | c2gwoC_ |
|
not modelled |
10.0 |
20 |
PDB header:hydrolase
Chain: C: PDB Molecule:dual specificity protein phosphatase 13;
PDBTitle: crystal structure of tmdp
|
| 45 | c3lcrA_ |
|
not modelled |
10.0 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:tautomycetin biosynthetic pks;
PDBTitle: thioesterase from tautomycetin biosynthhetic pathway
|
| 46 | d1jmxa1 |
|
not modelled |
9.5 |
15 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Quinohemoprotein amine dehydrogenase A chain, domains 1 and 2 |
| 47 | d1vhra_ |
|
not modelled |
8.4 |
17 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Dual specificity phosphatase-like |
| 48 | c2x5eA_ |
|
not modelled |
8.3 |
13 |
PDB header:unknown function
Chain: A: PDB Molecule:upf0271 protein pa4511;
PDBTitle: crystal structure of the hypothetical protein pa4511 from2 pseudomonas aeruginosa
|
| 49 | d1v6ta_ |
|
not modelled |
8.2 |
11 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:LamB/YcsF-like |
| 50 | d2dfaa1 |
|
not modelled |
7.9 |
10 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:LamB/YcsF-like |
| 51 | d2acya_ |
|
not modelled |
7.8 |
20 |
Fold:Ferredoxin-like
Superfamily:Acylphosphatase/BLUF domain-like
Family:Acylphosphatase-like |
| 52 | c2r37A_ |
|
not modelled |
7.7 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutathione peroxidase 3;
PDBTitle: crystal structure of human glutathione peroxidase 3 (selenocysteine to2 glycine mutant)
|
| 53 | d2ccaa2 |
|
not modelled |
7.7 |
15 |
Fold:Heme-dependent peroxidases
Superfamily:Heme-dependent peroxidases
Family:Catalase-peroxidase KatG |
| 54 | c3eurA_ |
|
not modelled |
7.1 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the c-terminal domain of uncharacterized protein2 from bacteroides fragilis nctc 9343
|
| 55 | d1cvla_ |
|
not modelled |
7.0 |
19 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Bacterial lipase |
| 56 | c3fleB_ |
|
not modelled |
6.9 |
12 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:se_1780 protein;
PDBTitle: se_1780 protein of unknown function from staphylococcus epidermidis.
|
| 57 | d1mwwa_ |
|
not modelled |
6.5 |
14 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:Hypothetical protein HI1388.1 |
| 58 | d3b5ea1 |
|
not modelled |
6.4 |
17 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Carboxylesterase/thioesterase 1 |
| 59 | d1pbya1 |
|
not modelled |
6.4 |
21 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Quinohemoprotein amine dehydrogenase A chain, domains 1 and 2 |
| 60 | d1gp1a_ |
|
not modelled |
6.3 |
36 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 61 | d2ccaa1 |
|
not modelled |
6.2 |
16 |
Fold:Heme-dependent peroxidases
Superfamily:Heme-dependent peroxidases
Family:Catalase-peroxidase KatG |
| 62 | c2p04B_ |
|
not modelled |
6.1 |
32 |
PDB header:transferase
Chain: B: PDB Molecule:signal transduction histidine kinase;
PDBTitle: 2.1 ang structure of the dimerized pas domain of signal transduction2 histidine kinase from nostoc punctiforme pcc 73102 with homology to3 the h-noxa/h-noba domain of the soluble guanylyl cyclase
|
| 63 | d1jfua_ |
|
not modelled |
6.0 |
18 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 64 | d1otfa_ |
|
not modelled |
5.9 |
17 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:4-oxalocrotonate tautomerase-like |
| 65 | d3buxb3 |
|
not modelled |
5.9 |
38 |
Fold:SH2-like
Superfamily:SH2 domain
Family:SH2 domain |
| 66 | d1xhoa_ |
|
not modelled |
5.8 |
4 |
Fold:Bacillus chorismate mutase-like
Superfamily:YjgF-like
Family:Chorismate mutase |
| 67 | c1xhoB_ |
|
not modelled |
5.8 |
4 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:chorismate mutase;
PDBTitle: chorismate mutase from clostridium thermocellum cth-682
|
| 68 | d1yt3a2 |
|
not modelled |
5.8 |
6 |
Fold:SAM domain-like
Superfamily:HRDC-like
Family:RNase D C-terminal domains |
| 69 | d1rbli_ |
|
not modelled |
5.7 |
14 |
Fold:RuBisCO, small subunit
Superfamily:RuBisCO, small subunit
Family:RuBisCO, small subunit |
| 70 | c3fi7A_ |
|
not modelled |
5.7 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:lmo1076 protein;
PDBTitle: crystal structure of the autolysin auto (lmo1076) from listeria2 monocytogenes, catalytic domain
|
| 71 | c1itkB_ |
|
not modelled |
5.6 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:catalase-peroxidase;
PDBTitle: crystal structure of catalase-peroxidase from haloarcula2 marismortui
|
| 72 | c3dwvB_ |
|
not modelled |
5.6 |
55 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glutathione peroxidase-like protein;
PDBTitle: glutathione peroxidase-type tryparedoxin peroxidase,2 oxidized form
|
| 73 | d2choa2 |
|
not modelled |
5.6 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:alpha-D-glucuronidase/Hyaluronidase catalytic domain |
| 74 | c2zyiB_ |
|
not modelled |
5.6 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:lipase, putative;
PDBTitle: a. fulgidus lipase with fatty acid fragment and calcium
|
| 75 | c2op8A_ |
|
not modelled |
5.5 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:probable tautomerase ywhb;
PDBTitle: crystal structure of ywhb- homologue of 4-oxalocrotonate tautomerase
|
| 76 | d2h1ia1 |
|
not modelled |
5.4 |
13 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Carboxylesterase/thioesterase 1 |
| 77 | c2e0tA_ |
|
not modelled |
5.4 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:dual specificity phosphatase 26;
PDBTitle: crystal structure of catalytic domain of dual specificity phosphatase2 26, ms0830 from homo sapiens
|
| 78 | d1svdm1 |
|
not modelled |
5.1 |
24 |
Fold:RuBisCO, small subunit
Superfamily:RuBisCO, small subunit
Family:RuBisCO, small subunit |
| 79 | d1uzhc1 |
|
not modelled |
5.1 |
10 |
Fold:RuBisCO, small subunit
Superfamily:RuBisCO, small subunit
Family:RuBisCO, small subunit |
| 80 | d2f8aa1 |
|
not modelled |
5.1 |
27 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 81 | c3hf2A_ |
|
not modelled |
5.1 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:bifunctional p-450/nadph-p450 reductase;
PDBTitle: crystal structure of the i401p mutant of cytochrome p450 bm3
|
| 82 | c1qgeD_ |
|
not modelled |
5.1 |
19 |
PDB header:hydrolase
Chain: D: PDB Molecule:protein (triacylglycerol hydrolase);
PDBTitle: new crystal form of pseudomonas glumae (formerly chromobacterium2 viscosum atcc 6918) lipase
|
| 83 | c2obiA_ |
|
not modelled |
5.1 |
36 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phospholipid hydroperoxide glutathione
PDBTitle: crystal structure of the selenocysteine to cysteine mutant2 of human phospholipid hydroperoxide glutathione peroxidase3 (gpx4)
|