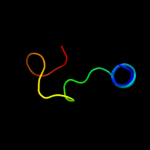
1 c3rceA_
96.5
14
PDB header: transferase/peptideChain: A: PDB Molecule: oligosaccharide transferase to n-glycosylate proteins;PDBTitle: bacterial oligosaccharyltransferase pglb
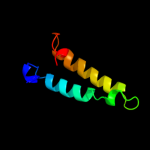
2 d1xapa_
23.7
13
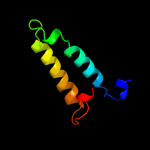
Fold: Nuclear receptor ligand-binding domainSuperfamily: Nuclear receptor ligand-binding domainFamily: Nuclear receptor ligand-binding domain3 c2pqfF_
19.9
17
PDB header: transferaseChain: F: PDB Molecule: poly [adp-ribose] polymerase 12;PDBTitle: human poly(adp-ribose) polymerase 12, catalytic fragment in complex2 with an inhibitor 3-aminobenzoic acid
4 d3d24a1
17.0
7
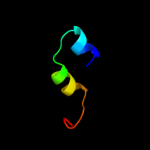
Fold: Nuclear receptor ligand-binding domainSuperfamily: Nuclear receptor ligand-binding domainFamily: Nuclear receptor ligand-binding domain5 c3d24A_
17.0
7
PDB header: transcriptionChain: A: PDB Molecule: steroid hormone receptor err1;PDBTitle: crystal structure of ligand-binding domain of estrogen-2 related receptor alpha (erralpha) in complex with the3 peroxisome proliferators-activated receptor coactivator-4 1alpha box3 peptide (pgc-1alpha)
6 c2pd0D_
15.9
47
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: hypothetical protein;PDBTitle: protein cgd2_2020 from cryptosporidium parvum
7 d1ef4a_
10.4
4
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: RNA polymerase subunit RPB10Family: RNA polymerase subunit RPB108 c2pmzN_
10.3
8
PDB header: translation, transferaseChain: N: PDB Molecule: dna-directed rna polymerase subunit n;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
9 c1yvyA_
9.9
21
PDB header: lyaseChain: A: PDB Molecule: phosphoenolpyruvate carboxykinase [atp];PDBTitle: crystal strucutre of anaerobiospirillum succiniciproducens2 phosphoenolpyruvate carboxykinase
10 d3pmga2
9.2
13
Fold: Phosphoglucomutase, first 3 domainsSuperfamily: Phosphoglucomutase, first 3 domainsFamily: Phosphoglucomutase, first 3 domains11 d1tuza_
9.2
36
Fold: EF Hand-likeSuperfamily: EF-handFamily: EF-hand modules in multidomain proteins12 d3eipa_
8.0
13
Fold: FKBP-likeSuperfamily: Colicin E3 immunity proteinFamily: Colicin E3 immunity protein13 d1a7ja_
7.8
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Phosphoribulokinase/pantothenate kinase14 c3do9C_
7.7
11
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: upf0302 protein ba_1542/gbaa1542/bas1430;PDBTitle: crystal structure of protein ba1542 from bacillus anthracis2 str.ames
15 d3bgea1
7.2
7
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: MgsA/YrvN C-terminal domain-like16 c2p0oA_
7.1
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein duf871;PDBTitle: crystal structure of a conserved protein from locus ef_2437 in2 enterococcus faecalis with an unknown function
17 c3rkoF_
7.1
17
PDB header: oxidoreductaseChain: F: PDB Molecule: nadh-quinone oxidoreductase subunit j;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
18 d1twfj_
7.0
7
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: RNA polymerase subunit RPB10Family: RNA polymerase subunit RPB1019 c3letB_
6.7
35
PDB header: transferaseChain: B: PDB Molecule: adenosine monophosphate-protein transferase vops;PDBTitle: crystal structure of fic domain containing ampylator, vops
20 c2ijoA_
6.7
9
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: polyprotein;PDBTitle: crystal structure of the west nile virus ns2b-ns3 protease2 complexed with bovine pancreatic trypsin inhibitor
21 d3ctda1
not modelled
6.3
7
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: MgsA/YrvN C-terminal domain-like22 c3e90C_
not modelled
6.2
9
PDB header: hydrolaseChain: C: PDB Molecule: ns2b cofactor;PDBTitle: west nile vi rus ns2b-ns3protease in complexed with2 inhibitor naph-kkr-h
23 d1osva_
not modelled
6.0
5
Fold: Nuclear receptor ligand-binding domainSuperfamily: Nuclear receptor ligand-binding domainFamily: Nuclear receptor ligand-binding domain24 c1x7fA_
not modelled
5.8
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: outer surface protein;PDBTitle: crystal structure of an uncharacterized b. cereus protein
25 d1x6aa2
not modelled
5.6
14
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain26 d2r9ga1
not modelled
5.5
14
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: MgsA/YrvN C-terminal domain-like27 c3et1A_
not modelled
5.4
15
PDB header: transcription/transferaseChain: A: PDB Molecule: peroxisome proliferator-activated receptor alpha;PDBTitle: structure of pparalpha with 3-[5-methoxy-1-(4-methoxy-2 benzenesulfonyl)-1h-indol-3-yl]-propionic acid