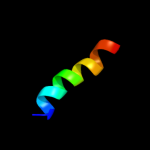
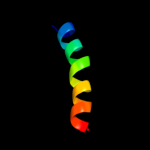
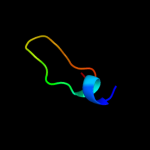
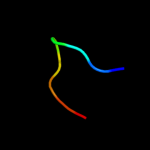
1 c2l3oA_
27.0
50
PDB header: cytokineChain: A: PDB Molecule: interleukin 3;PDBTitle: solution structure of murine interleukin 3
2 c2wwbB_
15.5
11
PDB header: ribosomeChain: B: PDB Molecule: protein transport protein sec61 subunit gamma;PDBTitle: cryo-em structure of the mammalian sec61 complex bound to the2 actively translating wheat germ 80s ribosome
3 d1rhzb_
13.0
14
Fold: Single transmembrane helixSuperfamily: Preprotein translocase SecE subunitFamily: Preprotein translocase SecE subunit4 c1h4uA_
12.1
11
PDB header: extracellular matrix proteinChain: A: PDB Molecule: nidogen-1;PDBTitle: domain g2 of mouse nidogen-1
5 d1gl4a1
10.2
11
Fold: GFP-likeSuperfamily: GFP-likeFamily: Domain G2 of nidogen-16 c3bhpA_
10.2
0
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0291 protein ynzc;PDBTitle: crystal structure of upf0291 protein ynzc from bacillus2 subtilis at resolution 2.0 a. northeast structural3 genomics consortium target sr384
7 c3kdvB_
9.1
89
PDB header: dna binding proteinChain: B: PDB Molecule: dna damage response b protein;PDBTitle: crystal structure of dna damage response b (ddrb) from deinococcus2 geothermalis
8 d1xrda1
8.9
18
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits9 d1wu9a1
8.8
6
Fold: EB1 dimerisation domain-likeSuperfamily: EB1 dimerisation domain-likeFamily: EB1 dimerisation domain-like10 c1ujlA_
8.6
18
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily hPDBTitle: solution structure of the herg k+ channel s5-p2 extracellular linker
11 d1txqb1
8.4
6
Fold: EB1 dimerisation domain-likeSuperfamily: EB1 dimerisation domain-likeFamily: EB1 dimerisation domain-like12 c1txqB_
8.4
6
PDB header: structural protein/protein bindingChain: B: PDB Molecule: microtubule-associated protein rp/eb familyPDBTitle: crystal structure of the eb1 c-terminal domain complexed2 with the cap-gly domain of p150glued
13 c2kncA_
8.4
18
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
14 c2qnaB_
8.0
15
PDB header: transport proteinChain: B: PDB Molecule: snurportin-1;PDBTitle: crystal structure of human importin-beta (127-876) in complex with the2 ibb-domain of snurportin1 (1-65)
15 c2latA_
7.6
36
PDB header: membrane proteinChain: A: PDB Molecule: dolichyl-diphosphooligosaccharide--proteinPDBTitle: solution structure of a human minimembrane protein ost4
16 c2ys9A_
7.6
33
PDB header: transcriptionChain: A: PDB Molecule: homeobox and leucine zipper protein homez;PDBTitle: structure of the third homeodomain from the human homeobox2 and leucine zipper protein, homez
17 c2eqoA_
7.4
27
PDB header: transcriptionChain: A: PDB Molecule: tnf receptor-associated factor 3-interactingPDBTitle: solution structure of the stn_traf3ip1_nd domain of2 interleukin 13 receptor alpha 1-binding protein-1 [homo3 sapiens]
18 c2gzdC_
7.3
18
PDB header: protein transportChain: C: PDB Molecule: rab11 family-interacting protein 2;PDBTitle: crystal structure of rab11 in complex with rab11-fip2
19 d2cpga_
7.3
40
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: CopG-like20 c2k1aA_
6.9
21
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
21 c3a98B_
not modelled
6.9
17
PDB header: signaling proteinChain: B: PDB Molecule: engulfment and cell motility protein 1;PDBTitle: crystal structure of the complex of the interacting regions of dock22 and elmo1
22 c3sojA_
not modelled
6.8
20
PDB header: cell adhesionChain: A: PDB Molecule: pile;PDBTitle: francisella tularensis pilin pile
23 c2qx7A_
not modelled
6.5
21
PDB header: plant proteinChain: A: PDB Molecule: eugenol synthase 1;PDBTitle: structure of eugenol synthase from ocimum basilicum
24 c2jvdA_
not modelled
6.4
0
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0291 protein ynzc;PDBTitle: solution nmr structure of the folded n-terminal fragment of2 upf0291 protein ynzc from bacillus subtilis. northeast3 structural genomics target sr384-1-46
25 c2wssT_
not modelled
6.4
14
PDB header: hydrolaseChain: T: PDB Molecule: atp synthase subunit b, mitochondrial;PDBTitle: the structure of the membrane extrinsic region of bovine2 atp synthase
26 d1szia_
not modelled
6.2
19
Fold: Four-helical up-and-down bundleSuperfamily: Mannose-6-phosphate receptor binding protein 1 (Tip47), C-terminal domainFamily: Mannose-6-phosphate receptor binding protein 1 (Tip47), C-terminal domain27 c2adlB_
not modelled
6.1
60
PDB header: dna binding proteinChain: B: PDB Molecule: ccda;PDBTitle: solution structure of the bacterial antitoxin ccda:2 implications for dna and toxin binding
28 c1jb0M_
not modelled
6.1
29
PDB header: photosynthesisChain: M: PDB Molecule: photosystem 1 reaction centre subunit xii;PDBTitle: crystal structure of photosystem i: a photosynthetic reaction center2 and core antenna system from cyanobacteria
29 d1jb0m_
not modelled
6.1
29
Fold: Single transmembrane helixSuperfamily: Subunit XII of photosystem I reaction centre, PsaMFamily: Subunit XII of photosystem I reaction centre, PsaM30 d1fc2c_
not modelled
6.0
30
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Bacterial immunoglobulin/albumin-binding domainsFamily: Immunoglobulin-binding protein A modules31 c2q5dD_
not modelled
5.9
10
PDB header: protein transportChain: D: PDB Molecule: snurportin-1;PDBTitle: crystal structure of human importin beta bound to the2 snurportin1 ibb-domain second crystal form
32 d1iq8a4
not modelled
5.8
20
Fold: Cystatin-likeSuperfamily: Pre-PUA domainFamily: Archaeosine tRNA-guanine transglycosylase, C2 domain33 c2k6sB_
not modelled
5.6
20
PDB header: protein transportChain: B: PDB Molecule: rab11fip2 protein;PDBTitle: structure of rab11-fip2 c-terminal coiled-coil domain
34 c3pvlB_
not modelled
5.6
45
PDB header: motor protein/protein transportChain: B: PDB Molecule: usher syndrome type-1g protein;PDBTitle: structure of myosin viia myth4-ferm-sh3 in complex with the cen1 of2 sans
35 d1nkzb_
not modelled
5.6
13
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits36 c2hg5D_
not modelled
5.6
13
PDB header: membrane proteinChain: D: PDB Molecule: kcsa channel;PDBTitle: cs+ complex of a k channel with an amide to ester substitution in the2 selectivity filter
37 c3mtuD_
not modelled
5.5
6
PDB header: contractile proteinChain: D: PDB Molecule: tropomyosin alpha-1 chain, microtubule-associated proteinPDBTitle: structure of the tropomyosin overlap complex from chicken smooth2 muscle
38 d1lp1b_
not modelled
5.5
30
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Bacterial immunoglobulin/albumin-binding domainsFamily: Immunoglobulin-binding protein A modules39 c2pbdV_
not modelled
5.5
71
PDB header: structural proteinChain: V: PDB Molecule: vasodilator-stimulated phosphoprotein;PDBTitle: ternary complex of profilin-actin with the poly-pro-gab2 domain of vasp*
40 c2ww9B_
not modelled
5.4
18
PDB header: ribosomeChain: B: PDB Molecule: protein transport protein sss1;PDBTitle: cryo-em structure of the active yeast ssh1 complex bound to the2 yeast 80s ribosome
41 d1prtf_
not modelled
5.3
21
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Bacterial AB5 toxins, B-subunits42 d2f2fa1
not modelled
5.3
33
Fold: beta-TrefoilSuperfamily: Ricin B-like lectinsFamily: Ricin B-like