| 1 |
|
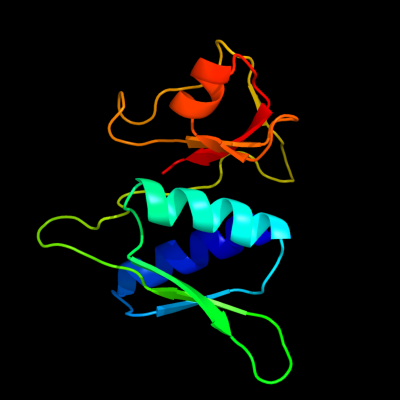
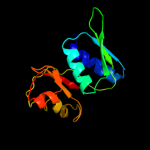
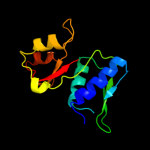
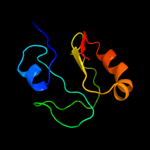
PDB 1sei chain A
Region: 3 - 129
Aligned: 127
Modelled: 127
Confidence: 100.0%
Identity: 51%
Fold: Ribosomal protein S8
Superfamily: Ribosomal protein S8
Family: Ribosomal protein S8
Phyre2
| 2 |
|
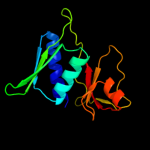
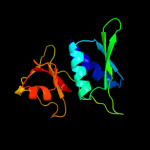
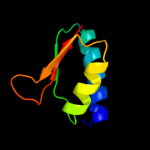
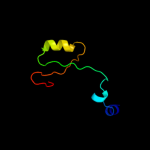
PDB 2gy9 chain H domain 1
Region: 4 - 129
Aligned: 126
Modelled: 126
Confidence: 100.0%
Identity: 100%
Fold: Ribosomal protein S8
Superfamily: Ribosomal protein S8
Family: Ribosomal protein S8
Phyre2
| 3 |
|
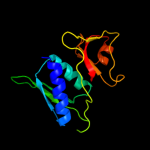
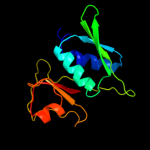
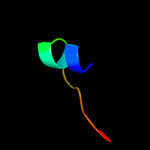
PDB 3rf2 chain A
Region: 4 - 129
Aligned: 126
Modelled: 126
Confidence: 100.0%
Identity: 50%
PDB header:ribosomal protein
Chain: A: PDB Molecule:30s ribosomal protein s8;
PDBTitle: crystal structure of 30s ribosomal protein s8 from aquifex aeolicus
Phyre2
| 4 |
|
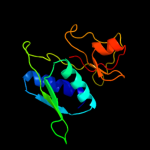
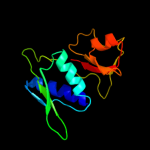
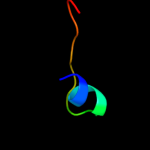
PDB 1i94 chain H
Region: 3 - 129
Aligned: 127
Modelled: 127
Confidence: 100.0%
Identity: 49%
Fold: Ribosomal protein S8
Superfamily: Ribosomal protein S8
Family: Ribosomal protein S8
Phyre2
| 5 |
|
PDB 1i6u chain A
Region: 2 - 129
Aligned: 125
Modelled: 128
Confidence: 100.0%
Identity: 28%
Fold: Ribosomal protein S8
Superfamily: Ribosomal protein S8
Family: Ribosomal protein S8
Phyre2
| 6 |
|
PDB 1an7 chain A
Region: 4 - 129
Aligned: 126
Modelled: 126
Confidence: 100.0%
Identity: 49%
Fold: Ribosomal protein S8
Superfamily: Ribosomal protein S8
Family: Ribosomal protein S8
Phyre2
| 7 |
|
PDB 3bbn chain H
Region: 1 - 129
Aligned: 128
Modelled: 129
Confidence: 100.0%
Identity: 45%
PDB header:ribosome
Chain: H: PDB Molecule:ribosomal protein s8;
PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
Phyre2
| 8 |
|
PDB 1s1h chain H
Region: 4 - 129
Aligned: 123
Modelled: 126
Confidence: 100.0%
Identity: 25%
PDB header:ribosome
Chain: H: PDB Molecule:40s ribosomal protein s22;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
Phyre2
| 9 |
|
PDB 2xzn chain H
Region: 2 - 129
Aligned: 125
Modelled: 128
Confidence: 100.0%
Identity: 22%
PDB header:ribosome
Chain: H: PDB Molecule:ribosomal protein s8 containing protein;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
Phyre2
| 10 |
|
PDB 2zqe chain A
Region: 1 - 64
Aligned: 64
Modelled: 64
Confidence: 25.2%
Identity: 13%
PDB header:dna binding protein
Chain: A: PDB Molecule:muts2 protein;
PDBTitle: crystal structure of the smr domain of thermus thermophilus muts2
Phyre2
| 11 |
|
PDB 2k19 chain A
Region: 5 - 46
Aligned: 42
Modelled: 42
Confidence: 17.0%
Identity: 14%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:putative piscicolin 126 immunity protein;
PDBTitle: nmr solution structure of pisi
Phyre2
| 12 |
|
PDB 2hgc chain A
Region: 37 - 54
Aligned: 18
Modelled: 18
Confidence: 16.5%
Identity: 39%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:yjcq protein;
PDBTitle: solution nmr structure of the yjcq protein from bacillus2 subtilis. northeast structural genomics target sr346.
Phyre2
| 13 |
|
PDB 2hgc chain A domain 1
Region: 37 - 54
Aligned: 18
Modelled: 18
Confidence: 16.5%
Identity: 39%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: YjcQ-like
Phyre2
| 14 |
|
PDB 2zrr chain A
Region: 4 - 46
Aligned: 43
Modelled: 43
Confidence: 15.3%
Identity: 16%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:mundticin ks immunity protein;
PDBTitle: crystal structure of an immunity protein that contributes2 to the self-protection of bacteriocin-producing3 enterococcus mundtii 15-1a
Phyre2
| 15 |
|
PDB 2dk5 chain A domain 1
Region: 63 - 87
Aligned: 25
Modelled: 25
Confidence: 10.7%
Identity: 20%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: RPO3F domain-like
Phyre2
| 16 |
|
PDB 1a6s chain A
Region: 28 - 48
Aligned: 21
Modelled: 21
Confidence: 10.5%
Identity: 33%
Fold: Retroviral matrix proteins
Superfamily: Retroviral matrix proteins
Family: GAG polyprotein M-domain
Phyre2
| 17 |
|
PDB 1khi chain A domain 2
Region: 1 - 27
Aligned: 26
Modelled: 27
Confidence: 10.3%
Identity: 12%
Fold: OB-fold
Superfamily: Nucleic acid-binding proteins
Family: Cold shock DNA-binding domain-like
Phyre2
| 18 |
|
PDB 2vkc chain A
Region: 30 - 64
Aligned: 31
Modelled: 35
Confidence: 7.8%
Identity: 26%
PDB header:hydrolase
Chain: A: PDB Molecule:nedd4-binding protein 2;
PDBTitle: solution structure of the b3bp smr domain
Phyre2
| 19 |
|
PDB 3ff4 chain A
Region: 30 - 126
Aligned: 72
Modelled: 76
Confidence: 7.6%
Identity: 21%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein chu_1412
Phyre2
| 20 |
|
PDB 2qlv chain F
Region: 3 - 60
Aligned: 57
Modelled: 58
Confidence: 7.4%
Identity: 14%
PDB header:transferase/protein binding
Chain: F: PDB Molecule:nuclear protein snf4;
PDBTitle: crystal structure of the heterotrimer core of the s.2 cerevisiae ampk homolog snf1
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|